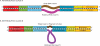A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing - PubMed (original) (raw)
A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing
Poornima Parameswaran et al. Nucleic Acids Res. 2007.
Abstract
Multiplexed high-throughput pyrosequencing is currently limited in complexity (number of samples sequenced in parallel), and in capacity (number of sequences obtained per sample). Physical-space segregation of the sequencing platform into a fixed number of channels allows limited multiplexing, but obscures available sequencing space. To overcome these limitations, we have devised a novel barcoding approach to allow for pooling and sequencing of DNA from independent samples, and to facilitate subsequent segregation of sequencing capacity. Forty-eight forward-reverse barcode pairs are described: each forward and each reverse barcode unique with respect to at least 4 nt positions. With improved read lengths of pyrosequencers, combinations of forward and reverse barcodes may be used to sequence from as many as n(2) independent libraries for each set of 'n' forward and 'n' reverse barcodes, for each defined set of cloning-linkers. In two pilot series of barcoded sequencing using the GS20 Sequencer (454/Roche), we found that over 99.8% of obtained sequences could be assigned to 25 independent, uniquely barcoded libraries based on the presence of either a perfect forward or a perfect reverse barcode. The false-discovery rate, as measured by the percentage of sequences with unexpected perfect pairings of unmatched forward and reverse barcodes, was estimated to be <0.005%.
Figures
Figure 1.
Design of forward and reverse primers. The synthesized primers are 45–46 nt long. (A and B) The template for the primers is: 454-Adapter:: Barcode:: Linker-Primer. Individual specifications for forward and reverse barcodes are also indicated. (C) Diagrammatic representation of the GS20 forward and reverse sequencing reads. ‘RC’ stands for Reverse Complement. Teeth denote base pairing.
Figure 2.
A diagrammatic representation of heteroduplexes that may be formed in an amplified pool. (A) Heteroduplexes formed between molecules from the same sample that have the same barcode, but different RNA inserts (X and Y). (B) Heteroduplexes formed between molecules with RNA inserts and molecules with no RNA inserts (or with fragments of linkers as inserts). Formation of these unusual duplexes may be facilitated by the 45–46 nt complementarity at either end of the insert. Thus, three types of molecules may be present during later stages of PCR: single-stranded, perfectly double stranded (Figure 1C) and heteroduplexes (shown here). The ratio of the three species is determined by the number of PCR cycles. As in Figure 1, ‘RC’ stands for Reverse Complement, and teeth denote base pairing.
Figure 3.
Visualizing the nature of amplified products from various cycles of PCR. With an increase in cycle number (twenty cycles of an initial round of PCR followed by six, eight or ten cycles of a second round of PCR), there is an evident shift in mobility of PCR products that contain a small RNA insert. Effect of PCR cycle number on three different samples is shown, stressing the importance of titrating the total number of DNA amplification cycles, to avoid saturation of the PCR amplification. Red arrows represent the two sizes of the insert-containing PCR products. PCR products without small RNA inserts migrate as faint bands between 75 and 100 bp (black arrow).
Similar articles
- Pair-barcode high-throughput sequencing for large-scale multiplexed sample analysis.
Tu J, Ge Q, Wang S, Wang L, Sun B, Yang Q, Bai Y, Lu Z. Tu J, et al. BMC Genomics. 2012 Jan 25;13:43. doi: 10.1186/1471-2164-13-43. BMC Genomics. 2012. PMID: 22276739 Free PMC article. - Impact of next-generation sequencing error on analysis of barcoded plasmid libraries of known complexity and sequence.
Deakin CT, Deakin JJ, Ginn SL, Young P, Humphreys D, Suter CM, Alexander IE, Hallwirth CV. Deakin CT, et al. Nucleic Acids Res. 2014;42(16):e129. doi: 10.1093/nar/gku607. Epub 2014 Jul 10. Nucleic Acids Res. 2014. PMID: 25013183 Free PMC article. - BARCOSEL: a tool for selecting an optimal barcode set for high-throughput sequencing.
Somervuo P, Koskinen P, Mei P, Holm L, Auvinen P, Paulin L. Somervuo P, et al. BMC Bioinformatics. 2018 Jul 5;19(1):257. doi: 10.1186/s12859-018-2262-7. BMC Bioinformatics. 2018. PMID: 29976145 Free PMC article. - Enhancing the detection of barcoded reads in high throughput DNA sequencing data by controlling the false discovery rate.
Buschmann T, Zhang R, Brash DE, Bystrykh LV. Buschmann T, et al. BMC Bioinformatics. 2014 Aug 7;15(1):264. doi: 10.1186/1471-2105-15-264. BMC Bioinformatics. 2014. PMID: 25099007 Free PMC article. - Dissecting intercellular and intracellular signaling networks with barcoded genetic tools.
Herholt A, Sahoo VK, Popovic L, Wehr MC, Rossner MJ. Herholt A, et al. Curr Opin Chem Biol. 2022 Feb;66:102091. doi: 10.1016/j.cbpa.2021.09.002. Epub 2021 Oct 10. Curr Opin Chem Biol. 2022. PMID: 34644670 Review.
Cited by
- Microbial shifts in the swine distal gut in response to the treatment with antimicrobial growth promoter, tylosin.
Kim HB, Borewicz K, White BA, Singer RS, Sreevatsan S, Tu ZJ, Isaacson RE. Kim HB, et al. Proc Natl Acad Sci U S A. 2012 Sep 18;109(38):15485-90. doi: 10.1073/pnas.1205147109. Epub 2012 Sep 6. Proc Natl Acad Sci U S A. 2012. PMID: 22955886 Free PMC article. - Rapid flow-sorting to simultaneously resolve multiplex massively parallel sequencing products.
Sandberg J, Werne B, Dessing M, Lundeberg J. Sandberg J, et al. Sci Rep. 2011;1:108. doi: 10.1038/srep00108. Epub 2011 Oct 6. Sci Rep. 2011. PMID: 22355625 Free PMC article. - Characterization of the oral fungal microbiome (mycobiome) in healthy individuals.
Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, Gillevet PM. Ghannoum MA, et al. PLoS Pathog. 2010 Jan 8;6(1):e1000713. doi: 10.1371/journal.ppat.1000713. PLoS Pathog. 2010. PMID: 20072605 Free PMC article. - A scalable, fully automated process for construction of sequence-ready barcoded libraries for 454.
Lennon NJ, Lintner RE, Anderson S, Alvarez P, Barry A, Brockman W, Daza R, Erlich RL, Giannoukos G, Green L, Hollinger A, Hoover CA, Jaffe DB, Juhn F, McCarthy D, Perrin D, Ponchner K, Powers TL, Rizzolo K, Robbins D, Ryan E, Russ C, Sparrow T, Stalker J, Steelman S, Weiand M, Zimmer A, Henn MR, Nusbaum C, Nicol R. Lennon NJ, et al. Genome Biol. 2010;11(2):R15. doi: 10.1186/gb-2010-11-2-r15. Epub 2010 Feb 5. Genome Biol. 2010. PMID: 20137071 Free PMC article. - The marriage of nutrigenomics with the microbiome: the case of infant-associated bifidobacteria and milk.
Sela DA, Mills DA. Sela DA, et al. Am J Clin Nutr. 2014 Mar;99(3):697S-703S. doi: 10.3945/ajcn.113.071795. Epub 2014 Jan 22. Am J Clin Nutr. 2014. PMID: 24452239 Free PMC article.
References
- Bennett ST, Barnes C, Cox A, Davies L, Brown C. Toward the 1,000 dollars human genome. Pharmacogenomics. 2005;6:373–382. - PubMed
- Ahmadian A, Ehn M, Hober S. Pyrosequencing: history, biochemistry and future. Clin. Chim. Acta. 2006;363:83–94. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HG003571/HG/NHGRI NIH HHS/United States
- 1S10RR022982/RR/NCRR NIH HHS/United States
- R01HG003571/HG/NHGRI NIH HHS/United States
- R01 GM37706/GM/NIGMS NIH HHS/United States
- R01 GM037706/GM/NIGMS NIH HHS/United States
- S10 RR022982/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials