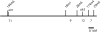Differential effect of allorecognition loci on phenotype in Hydractinia symbiolongicarpus (Cnidaria: Hydrozoa) - PubMed (original) (raw)
Differential effect of allorecognition loci on phenotype in Hydractinia symbiolongicarpus (Cnidaria: Hydrozoa)
Anahid E Powell et al. Genetics. 2007 Dec.
Abstract
The allorecognition complex of Hydractinia symbiolongicarpus is a chromosomal interval containing two loci, alr1 and alr2, that controls fusion between genetically distinct colonies. Recombination between these two loci has been associated with a heterogeneous class of phenotypes called transitory fusion. A large-scale backcross was performed to generate a population of colonies (N = 106) with recombination breakpoints within the allorecognition complex. Two distinct forms of transitory fusion were correlated with reciprocal recombination products, suggesting that alr1 and alr2 contributed differentially to the allorecognition response. Specifically, type I transitory fusion is associated with rapid and persistent separation of allogeneic tissues, whereas type II transitory fusion generates a patchwork of continuously fusing and separating tissues.
Figures
Figure 1.—
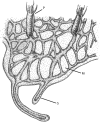
Schematic showing the morphology of a Hydractinia colony. P, polyp; M, mat; S, stolon.
Figure 2.—
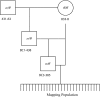
Pedigree used to generate mapping population. 431-63 is a heterozygote, and 833-8 is a homozygote. Squares represent males, and circles represent females. The lineage of 833-8 can be found in Figure 2 of C
adavid
et al. (2004) and likewise the heterozygous male (431-63) was a member of the 431 population represented as the final product of the mating program in C
adavid
et al. (2004).
Figure 3.—
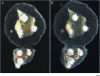
Initial stages of fusion and transitory fusion. (A) Colony assay prior to contact where the two colonies grow side by side. (B) Fusion 48 hr later showing continuity between mat tissue and gastrovascular canals. Bar, 200 μm.
Figure 4.—
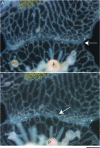
Outcome of type I transitory fusion. This close-up of interacting tissue between colonies depicts the outcome of type I transitory fusion. The two colonies have already undergone fusion and initial stages of rejection and have separated, indicated by the white fibrous material between the colonies that creates a border (arrow). Note also the gastrovascular canals that branch laterally along the line of separation and rejoin one another. P, polyps. Bar, 200 μm.
Figure 5.—
Two stages of type II transitory fusion. This region of interacting tissue between two colonies depicts the separation and refusion phases that occur in type II transitory fusion. (A) The separation phase after the colony has already undergone contact and fusion. The zone of interaction/separation is indicated by an arrow. This is an early stage of separation common to both type I and II transitory fusion where the mat and gastrovascular canals are separate, but unlike Figure 4, there is no border accumulation or extensive lateral branching of canals. (B) Refusion 24 hr later. The arrow indicates one of several areas where colonies have rejoined their gastrovascular canals. P, polyps. Bar, 200 μm.
Figure 6.—
Genetic map of the allorecognition complex. Markers are named as in Table 1. Loci are alr1 and alr2. Bar, 0.1 cM as shown. The total mapped interval represents 1.7 cM. Numbers below the map represent how many recombinants were recovered between each of the markers.
Similar articles
- The Hydractinia allorecognition system.
Nicotra ML. Nicotra ML. Immunogenetics. 2022 Feb;74(1):27-34. doi: 10.1007/s00251-021-01233-6. Epub 2021 Nov 13. Immunogenetics. 2022. PMID: 34773127 Review. - Multiple Alr genes exhibit allorecognition-associated variation in the colonial cnidarian Hydractinia.
Rodriguez-Valbuena H, Gonzalez-Muñoz A, Cadavid LF. Rodriguez-Valbuena H, et al. Immunogenetics. 2022 Dec;74(6):559-581. doi: 10.1007/s00251-022-01268-3. Epub 2022 Jun 27. Immunogenetics. 2022. PMID: 35761101 - The putative immune recognition repertoire of the model cnidarian Hydractinia symbiolongicarpus is large and diverse.
Zárate-Potes A, Ocampo ID, Cadavid LF. Zárate-Potes A, et al. Gene. 2019 Feb 5;684:104-117. doi: 10.1016/j.gene.2018.10.068. Epub 2018 Oct 26. Gene. 2019. PMID: 30393111 - Allorecognition proteins in an invertebrate exhibit homophilic interactions.
Karadge UB, Gosto M, Nicotra ML. Karadge UB, et al. Curr Biol. 2015 Nov 2;25(21):2845-2850. doi: 10.1016/j.cub.2015.09.030. Epub 2015 Oct 8. Curr Biol. 2015. PMID: 26455308 Free PMC article. - Model systems of invertebrate allorecognition.
Rosengarten RD, Nicotra ML. Rosengarten RD, et al. Curr Biol. 2011 Jan 25;21(2):R82-92. doi: 10.1016/j.cub.2010.11.061. Curr Biol. 2011. PMID: 21256442 Review.
Cited by
- The Hydractinia allorecognition system.
Nicotra ML. Nicotra ML. Immunogenetics. 2022 Feb;74(1):27-34. doi: 10.1007/s00251-021-01233-6. Epub 2021 Nov 13. Immunogenetics. 2022. PMID: 34773127 Review. - Origin and Evolution of the Sponge Aggregation Factor Gene Family.
Grice LF, Gauthier MEA, Roper KE, Fernàndez-Busquets X, Degnan SM, Degnan BM. Grice LF, et al. Mol Biol Evol. 2017 May 1;34(5):1083-1099. doi: 10.1093/molbev/msx058. Mol Biol Evol. 2017. PMID: 28104746 Free PMC article. - Invertebrate allorecognition: the origins of histocompatibility.
Dishaw LJ, Litman GW. Dishaw LJ, et al. Curr Biol. 2009 Apr 14;19(7):R286-8. doi: 10.1016/j.cub.2009.02.035. Curr Biol. 2009. PMID: 19368870 Free PMC article. - Hydractinia allodeterminant alr1 resides in an immunoglobulin superfamily-like gene complex.
Rosa SF, Powell AE, Rosengarten RD, Nicotra ML, Moreno MA, Grimwood J, Lakkis FG, Dellaporta SL, Buss LW. Rosa SF, et al. Curr Biol. 2010 Jun 22;20(12):1122-7. doi: 10.1016/j.cub.2010.04.050. Epub 2010 May 27. Curr Biol. 2010. PMID: 20537535 Free PMC article. - High fusibility and chimera prevalence in an invasive colonial ascidian.
Casso M, Tagliapietra D, Turon X, Pascual M. Casso M, et al. Sci Rep. 2019 Oct 30;9(1):15673. doi: 10.1038/s41598-019-51950-y. Sci Rep. 2019. PMID: 31666562 Free PMC article.
References
- Blackstone, N. W., and L. W. Buss, 1991. Shape variation in hydractiniid hydroids. Biol. Bull. 180: 394–405. - PubMed
- Buss, L. W., and R. K. Grosberg, 1990. Morphogenetic basis for phenotypic differences in hydroid competitive behavior. Nature 343: 63–66.
- Buss, L. W., C. S. McFadden and D. R. Keene, 1984. Biology of hydractiniid hydroids. 2. Histocompatibility effector system/competitive mechanism mediated by nematocyst discharge. Biol. Bull. 167: 139–158.
Publication types
MeSH terms
Grants and funding
- R21 AI066242/AI/NIAID NIH HHS/United States
- T32 GM007499/GM/NIGMS NIH HHS/United States
- T32-GM07499/GM/NIGMS NIH HHS/United States
- 1R21-AI066242/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials