The Alzheimer's disease beta-secretase enzyme, BACE1 - PubMed (original) (raw)
The Alzheimer's disease beta-secretase enzyme, BACE1
Sarah L Cole et al. Mol Neurodegener. 2007.
Abstract
The pathogenesis of Alzheimer's disease is highly complex. While several pathologies characterize this disease, amyloid plaques, composed of the beta-amyloid peptide are hallmark neuropathological lesions in Alzheimer's disease brain. Indeed, a wealth of evidence suggests that beta-amyloid is central to the pathophysiology of AD and is likely to play an early role in this intractable neurodegenerative disorder. The BACE1 enzyme is essential for the generation of beta-amyloid. BACE1 knockout mice do not produce beta-amyloid and are free from Alzheimer's associated pathologies including neuronal loss and certain memory deficits. The fact that BACE1 initiates the formation of beta-amyloid, and the observation that BACE1 levels are elevated in this disease provide direct and compelling reasons to develop therapies directed at BACE1 inhibition thus reducing beta-amyloid and its associated toxicities. However, new data indicates that complete abolishment of BACE1 may be associated with specific behavioral and physiological alterations. Recently a number of non-APP BACE1 substrates have been identified. It is plausible that failure to process certain BACE1 substrates may underlie some of the reported abnormalities in the BACE1-deficient mice. Here we review BACE1 biology, covering aspects ranging from the initial identification and characterization of this enzyme to recent data detailing the apparent dysregulation of BACE1 in Alzheimer's disease. We pay special attention to the putative function of BACE1 during healthy conditions and discuss in detail the relationship that exists between key risk factors for AD, such as vascular disease (and downstream cellular consequences), and the pathogenic alterations in BACE1 that are observed in the diseased state.
Figures
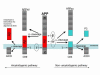
Figure 1
APP metabolism by the secretase enzymes. APP is sequentially cleaved by BACE1, the β-secretase, and γ-secretase, a complex comprised of presenilin, nicastin, Aph1 and Pen2, to generate Aβ. BACE1 cleavage of APP is a prerequisite for Aβ formation and is putatively the rate-limiting step in Aβ genesis. BACE1 cleavage of APP forms the N-terminus of the peptide, and two cleavage fragments are liberated: APPsβ, a secreted ectodomain, and C99, a membrane bound fragment. C99 is the substrate for γ-secretase, and C99 cleavage generates the AICD together with the C-terminus of Aβ. Aβ formation is prevented by the activities of α-secretase, which has been identified as TACE, ADAM9 and ADAM10. α-secretase cleaves APP to generate the secreted ectodomain, APPsα and membrane bound fragment, C83. C83 is subsequently cleaved by the γ-secretase complex to yield the 3 KDa fragment, P3 and the AICD.
Figure 2
The intracellular trafficking of BACE1. (1) Following synthesis in the ER, pro-BACE1 traffics to the Golgi. Here, the propeptide is removed and BACE1 undergoes significant post-translational modifications. The trafficking and localization of BACE1 appear dependent on the ACDL motif, composed of DISLL residues, in its C-terminal tail. Importantly, phosphorylation of the serine 498 residue (S498) is important for binding of BACE1 to the GGA monomeric adaptor proteins which are implicated in the sorting of cargo between the TGN and endosomes and vice versa [127-131], in addition to the Golgi complex to the endosomes [130]. (2) BACE1 traffics to the plasma membrane and while the re-internalization of BACE1 from the cell surface appears to be independent of the phosphorylation state of S498 [131], it has been reported that the di-leucine residues are important for BACE1 endocytosis [126]. Non-phosphorylated BACE1 may recycle from the endosome back to the cell surface [131]. However, BACE1 phosphorylated at the S498 site interacts with GGA1 in the endosome and traffics back to the TGN [131], and (3) the interaction of phosphorylated BACE1 with GGA3 appears to direct BACE1 to the lysosome, where it is degraded [135]. In addition to S498 phosphorylation, the di-leucine residues are also important for the sorting of BACE1 into lysosomal compartments [140]. BACE1 is also degraded in the proteasome [133]. The precise subcellular localization (early verses late endosomes) of the BACE1-GGA complex requires determination, and while it appears likely that BACE1 can traffic from the TGN to the endosome directly, it remains to be demonstrated experimentally.
Similar articles
- BACE1 structure and function in health and Alzheimer's disease.
Cole SL, Vassar R. Cole SL, et al. Curr Alzheimer Res. 2008 Apr;5(2):100-20. doi: 10.2174/156720508783954758. Curr Alzheimer Res. 2008. PMID: 18393796 Review. - The Basic Biology of BACE1: A Key Therapeutic Target for Alzheimer's Disease.
Cole SL, Vassar R. Cole SL, et al. Curr Genomics. 2007 Dec;8(8):509-30. doi: 10.2174/138920207783769512. Curr Genomics. 2007. PMID: 19415126 Free PMC article. - Review of synthesis, biological assay and QSAR studies of β-secretase inhibitors.
Niño H, García-Pintos I, Rodríguez-Borges JE, Escobar-Cubiella M, García-Mera X, Prado-Prado F. Niño H, et al. Curr Comput Aided Drug Des. 2011 Dec;7(4):263-75. doi: 10.2174/157340911798260322. Curr Comput Aided Drug Des. 2011. PMID: 22050682 Review. - The beta-secretase, BACE: a prime drug target for Alzheimer's disease.
Vassar R. Vassar R. J Mol Neurosci. 2001 Oct;17(2):157-70. doi: 10.1385/JMN:17:2:157. J Mol Neurosci. 2001. PMID: 11816789 Review. - beta-Secretase, APP and Abeta in Alzheimer's disease.
Vassar R. Vassar R. Subcell Biochem. 2005;38:79-103. Subcell Biochem. 2005. PMID: 15709474 Review.
Cited by
- Inhibition of γ-secretase worsens memory deficits in a genetically congruous mouse model of Danish dementia.
Tamayev R, D'Adamio L. Tamayev R, et al. Mol Neurodegener. 2012 Apr 26;7:19. doi: 10.1186/1750-1326-7-19. Mol Neurodegener. 2012. PMID: 22537414 Free PMC article. - Apolipoprotein E4 (1-272) fragment is associated with mitochondrial proteins and affects mitochondrial function in neuronal cells.
Nakamura T, Watanabe A, Fujino T, Hosono T, Michikawa M. Nakamura T, et al. Mol Neurodegener. 2009 Aug 20;4:35. doi: 10.1186/1750-1326-4-35. Mol Neurodegener. 2009. PMID: 19695092 Free PMC article. - Beta-Secretase 1 Recruits Amyloid-Beta Precursor Protein to ROCK2 Kinase, Resulting in Erroneous Phosphorylation and Beta-Amyloid Plaque Formation.
Hajdú I, Végh BM, Szilágyi A, Závodszky P. Hajdú I, et al. Int J Mol Sci. 2023 Jun 21;24(13):10416. doi: 10.3390/ijms241310416. Int J Mol Sci. 2023. PMID: 37445593 Free PMC article. - Scaffold Morphing and In Silico Design of Potential BACE-1 (β-Secretase) Inhibitors: A Hope for a Newer Dawn in Anti-Alzheimer Therapeutics.
Bhatia S, Singh M, Sharma P, Mujwar S, Singh V, Mishra KK, Singh TG, Singh T, Ahmad SF. Bhatia S, et al. Molecules. 2023 Aug 12;28(16):6032. doi: 10.3390/molecules28166032. Molecules. 2023. PMID: 37630283 Free PMC article. - Inclusion body myositis: a view from the Caenorhabditis elegans muscle.
Rebolledo DL, Minniti AN, Grez PM, Fadic R, Kohn R, Inestrosa NC. Rebolledo DL, et al. Mol Neurobiol. 2008 Oct;38(2):178-98. doi: 10.1007/s12035-008-8041-0. Epub 2008 Sep 5. Mol Neurobiol. 2008. PMID: 18773311 Review.
References
- Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L, Mant R, Newton P, Rooke K, Roques P, Talbot C, Pericak-Vance M, Roses A, Williamson R, Rossor M, Owen M, Hardy J. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. - DOI - PubMed
- St. George-Hyslop PH, Haines JL, Farrer LA, Polinsky R, Van Broeckhoven C, Goate A, Crapper McLachlan DR, Orr H, Bruni AC, Sorbi S, Rainero I, Foncin JF, Pollen D, Cantu JM, Tupler R, Voskresenskaya N, Mayeux R, Growdon J, Fried VA, Myers RH, Nee L, Backhovens H, Martin JJ, Rossor M, Owen MJ, Mullan M, Percy ME, Karlinsky H, Rich S, Heston L, Montesi M, Mortilla M, Nacmias N, Gusella JF, Hardy JA. Genetic linkage studies suggest that Alzheimer's disease is not a single homogeneous disorder. Nature. 1990;347:194–197. doi: 10.1038/347194a0. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources