Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase - PubMed (original) (raw)
. 2007 Dec 7;318(5856):1637-40.
doi: 10.1126/science.1150034. Epub 2007 Nov 15.
J Ross Chapman, Shinichiro Nakada, Jarkko Ylanko, Richard Chahwan, Frédéric D Sweeney, Stephanie Panier, Megan Mendez, Jan Wildenhain, Timothy M Thomson, Laurence Pelletier, Stephen P Jackson, Daniel Durocher
Affiliations
- PMID: 18006705
- PMCID: PMC2430610
- DOI: 10.1126/science.1150034
Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase
Nadine K Kolas et al. Science. 2007.
Expression of concern in
- Editorial Expression of Concern.
Thorp HH. Thorp HH. Science. 2024 Dec 6;386(6726):1102. doi: 10.1126/science.adu7744. Epub 2024 Dec 5. Science. 2024. PMID: 39637007 No abstract available.
Abstract
Cells respond to DNA double-strand breaks by recruiting factors such as the DNA-damage mediator protein MDC1, the p53-binding protein 1 (53BP1), and the breast cancer susceptibility protein BRCA1 to sites of damaged DNA. Here, we reveal that the ubiquitin ligase RNF8 mediates ubiquitin conjugation and 53BP1 and BRCA1 focal accumulation at sites of DNA lesions. Moreover, we establish that MDC1 recruits RNF8 through phosphodependent interactions between the RNF8 forkhead-associated domain and motifs in MDC1 that are phosphorylated by the DNA-damage activated protein kinase ataxia telangiectasia mutated (ATM). We also show that depletion of the E2 enzyme UBC13 impairs 53BP1 recruitment to sites of damage, which suggests that it cooperates with RNF8. Finally, we reveal that RNF8 promotes the G2/M DNA damage checkpoint and resistance to ionizing radiation. These results demonstrate how the DNA-damage response is orchestrated by ATM-dependent phosphorylation of MDC1 and RNF8-mediated ubiquitination.
Figures
Fig. 1
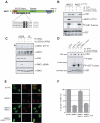
The MDC1 TQXF motifs are ATM targets required for 53BP1 IRIF. (A) Domain architecture of MDC1, with ATM consensus sites (dots). (B) MDC1 T719 is phosphorylated by ATM in vitro. GST-MDC1679-778 (fMDC1) or GST-MDC1679-778-T719A (fMDC1T719A) were incubated with anti-ATM or pre-immune complexes in the presence or absence of the ATM kinase inhibitor KU55933. (C) MDC1 TQXF motifs are phosphorylated by ATM in vivo. Lysates from U2OS or AT22IJE (AT) cells were immunoblotted with the indicated antibodies. The arrowhead points to phospho-MDC1. (D) The MDC1 TQXF motifs are phosphorylated by ATM in vitro. Kinase reactions with anti-ATM or pre-immune complexes and the following substrates: GST, GST-p531-56 (p53), fMDC1 or GST-MDC1679-778-AQXF (fMDC1AQXF) as in (B). (E-F) The TQXF cluster is required for 53BP1 IRIF. U2OS cells expressing siRNA-resistant GFP-MDC1 (MDC1*) or GFP-MDC1AQXF (MDC1*AQXF) were transfected with siRNA against MDC1 (siMDC1) or luciferase (siCTRL) and post-irradiation (5 Gy) were stained for MDC1 and 53BP1 (E) and quantitated (F; N=4 +/-SD).
Fig. 2
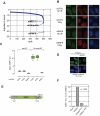
RNF8 promotes 53BP1 IRIF assembly. (A) Ranking by z-score of 500 siRNAs giving the least 53BP1 foci from an ongoing siRNA screen examining 53BP1 focus formation. See Fig. S5 for details. (B) 53BP1 and MDC1 immunofluorescence of U2OS cells transfected with control siRNA (siCTRL) or RNF8 siRNA (siRNF8) and fixed prior (no IR) or 1 hr following 10 Gy irradiation. (C) Quantitation of 53BP1 IRIF in HeLa cells transfected with the indicated esiRNAs (CTRL against luciferase). N=16, data displayed using box-and-whisker plots. (D) Transfection of siRNAi-resistant murine RNF8 in HeLa cells restores 53BP1 IRIF formation caused by RNF8 depletion. (E) Domain architecture of RNF8. Numbering refers to murine RNF8. (F) Rescue of RNF8 depletion by murine RNF8 but not the FHA- (R42A) or RING finger-mutated (C406) mutants. 53BP1 foci were quantitated 1 hr after 10 Gy irradiation in siRNF8-treated cells. Data for wild-type RNF8 were set at 100%. Over 1400 cells per condition were counted.
Fig. 3
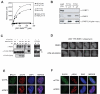
RNF8 mediates BRCA1-RAP80 IRIF via a physical interaction with MDC1. (A) Binding curves of GST-RNF8FHA (WT) the R42A mutant obtained by surface plasmon resonance with peptides corresponding to MDC1 epitopes surrounding T719, T752 or their phosphorylated counterparts (pT719 and pT752). (B) Peptide pull-downs of HeLa nuclear extracts with immobilized peptides; phosphorylated (pT) or unphosphorylated (T), encompassing MDC1 T719 or T752 residues. (C) RNF8 interacts with MDC1 in vivo. Extracts from 293T cells mock-transfected (-) or transfected with RNF8-FLAG (WT) or the R42A FHA mutant were immunoprecipitated (IP) with anti-MDC1 (M), anti-FLAG (F) or normal mouse IgG and probed for MDC1 or RNF8-FLAG as indicated. Arrowheads indicate the RNF8-specific signal. Note that RNF8 appears modified when interacting with MDC1. HC, IgG heavy chains. (D) Time-lapse microscopy of 293T cells stably expressing YFP-RNF8 pre-incubated with ATM inhibitor KU55933 or DMSO and treated with the radio-mimetic drug phleomycin (1.5 mg/ml) for the indicated times (min:sec). Three-dimensional image datasets were computationally deconvolved and shown as two-dimensional projections. Scale bar = 10μm. (E-F) Irradiated (10 Gy) HeLa cells transfected with the indicated siRNAs were stained with anti-γH2AX, anti-BRCA1 (D) or anti-RAP80 (E) antibodies 1hr post-IR.
Fig. 4
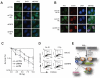
RNF8 cooperates with UBC13 to mediate ubiquitin IRIF and a functional DDR. (A-B) Irradiated (10 Gy) U2OS cells transfected with the indicated siRNA (A) or esiRNAs (B) were stained with FK2 anti- conjugated ubiquitin and γH2AX antibodies to assess IRIF. Cells were fixed 1 hr post-IR. (C) Clonogenic survival of HeLa cells transfected with siRNAs against ATM (siATM), RNF8 (siRNF8) or a non-targeting control (siCTRL). N=3 +/- SEM. (D) G2/M checkpoint analysis of U2OS cells transfected with the indicated esiRNAs. Fixed mock-treated (No IR) or irradiated (2 Gy) cells were stained with an anti-phospho-histone H3 antibody and propidium iodide (PI). The percentage of mitotic cells was determined by fluorescence-activated cell sorting. (E) Model of RNF8 action at DSBs. RNF8 is recruited by ATM-phosphorylated MDC1 to DSBs where it ubiquitinates an unknown protein (X), recruiting RAP80-BRCA1 and allowing 53BP1 to recognize methylated histones, as suggested by the RNF8-dependent recruitment of the 53BP1 Tudor domain to DSB sites (Fig. S13). Our results also suggest the presence of a MDC1-independent pathway of RNF8 action (dashed line) that mediate 53BP1 and BRCA1 IRIF formation.
Similar articles
- ATM activation accompanies histone H2AX phosphorylation in A549 cells upon exposure to tobacco smoke.
Tanaka T, Huang X, Jorgensen E, Gietl D, Traganos F, Darzynkiewicz Z, Albino AP. Tanaka T, et al. BMC Cell Biol. 2007 Jun 26;8:26. doi: 10.1186/1471-2121-8-26. BMC Cell Biol. 2007. PMID: 17594478 Free PMC article. - The radiomimetic enediyne C-1027 induces unusual DNA damage responses to double-strand breaks.
Kennedy DR, Beerman TA. Kennedy DR, et al. Biochemistry. 2006 Mar 21;45(11):3747-54. doi: 10.1021/bi052334c. Biochemistry. 2006. PMID: 16533058 Free PMC article. - PCNA-binding activity separates RNF168 functions in DNA replication and DNA double-stranded break signaling.
Yang Y, Jayaprakash D, Jhujh SS, Reynolds JJ, Chen S, Gao Y, Anand JR, Mutter-Rottmayer E, Ariel P, An J, Cheng X, Pearce KH, Blanchet SA, Nandakumar N, Zhou P, Fradet-Turcotte A, Stewart GS, Vaziri C. Yang Y, et al. Nucleic Acids Res. 2024 Nov 27;52(21):13019-13035. doi: 10.1093/nar/gkae918. Nucleic Acids Res. 2024. PMID: 39445802 Free PMC article. - Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - Pharmacological treatments in panic disorder in adults: a network meta-analysis.
Guaiana G, Meader N, Barbui C, Davies SJ, Furukawa TA, Imai H, Dias S, Caldwell DM, Koesters M, Tajika A, Bighelli I, Pompoli A, Cipriani A, Dawson S, Robertson L. Guaiana G, et al. Cochrane Database Syst Rev. 2023 Nov 28;11(11):CD012729. doi: 10.1002/14651858.CD012729.pub3. Cochrane Database Syst Rev. 2023. PMID: 38014714 Free PMC article. Review.
Cited by
- Molecular insights into the function of RING finger (RNF)-containing proteins hRNF8 and hRNF168 in Ubc13/Mms2-dependent ubiquitylation.
Campbell SJ, Edwards RA, Leung CC, Neculai D, Hodge CD, Dhe-Paganon S, Glover JN. Campbell SJ, et al. J Biol Chem. 2012 Jul 6;287(28):23900-10. doi: 10.1074/jbc.M112.359653. Epub 2012 May 15. J Biol Chem. 2012. PMID: 22589545 Free PMC article. - Impact of histone H4 lysine 20 methylation on 53BP1 responses to chromosomal double strand breaks.
Hartlerode AJ, Guan Y, Rajendran A, Ura K, Schotta G, Xie A, Shah JV, Scully R. Hartlerode AJ, et al. PLoS One. 2012;7(11):e49211. doi: 10.1371/journal.pone.0049211. Epub 2012 Nov 28. PLoS One. 2012. PMID: 23209566 Free PMC article. - CBX4-mediated SUMO modification regulates BMI1 recruitment at sites of DNA damage.
Ismail IH, Gagné JP, Caron MC, McDonald D, Xu Z, Masson JY, Poirier GG, Hendzel MJ. Ismail IH, et al. Nucleic Acids Res. 2012 Jul;40(12):5497-510. doi: 10.1093/nar/gks222. Epub 2012 Mar 8. Nucleic Acids Res. 2012. PMID: 22402492 Free PMC article. - RNF8 regulates active epigenetic modifications and escape gene activation from inactive sex chromosomes in post-meiotic spermatids.
Sin HS, Barski A, Zhang F, Kartashov AV, Nussenzweig A, Chen J, Andreassen PR, Namekawa SH. Sin HS, et al. Genes Dev. 2012 Dec 15;26(24):2737-48. doi: 10.1101/gad.202713.112. Genes Dev. 2012. PMID: 23249736 Free PMC article. - Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation.
Li M, Yu X. Li M, et al. Cancer Cell. 2013 May 13;23(5):693-704. doi: 10.1016/j.ccr.2013.03.025. Cancer Cell. 2013. PMID: 23680151 Free PMC article.
References
- Bartek J, Lukas J. Curr Opin Cell Biol. 2007;19:238. - PubMed
- Stiff T, et al. Cancer Res. 2004;64:2390. - PubMed
- Paull TT, et al. Curr Biol. 2000;10:886. - PubMed
- Celeste A, et al. Nat Cell Biol. 2003;5:675. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous