ID genes mediate tumor reinitiation during breast cancer lung metastasis - PubMed (original) (raw)
ID genes mediate tumor reinitiation during breast cancer lung metastasis
Gaorav P Gupta et al. Proc Natl Acad Sci U S A. 2007.
Abstract
The establishment of distant metastases depends on the capacity of small numbers of cancer cells to regenerate a tumor after entering a target tissue. The mechanisms that confer this capacity remain to be defined. Here we identify a role for the transcriptional inhibitors of differentiation Id1 and Id3 as selective mediators of lung metastatic colonization in the triple negative [TN, i.e., lacking expression of estrogen receptor and progesterone receptor, and lacking Her2 (human epidermal growth factor receptor 2) amplification] subgroup of human breast cancer. Although broad expression of Id1 has recently been documented in tumors of the rare metaplastic subtype, here we report that rare Id1-expressing cells are also present in the more common TN subset of human breast tumors but not in other subtypes. We also provide evidence that Id1 expression is enriched in clinically obtained hormone receptor negative lung metastases. Functional studies demonstrate that Id1 and its closely related family member Id3 are required for tumor initiating functions, both in the context of primary tumor formation and during metastatic colonization of the lung microenvironment. In vivo characterization of lung metastatic progression reveals that Id1 and Id3 facilitate sustained proliferation during the early stages of metastatic colonization, subsequent to extravasation into the lung parenchyma. These results shed light on the proliferative mechanisms that initiate metastatic colonization, and they implicate Id1 and Id3 as mediators of this malignant function in the TN subgroup of breast cancers.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
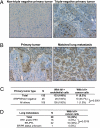
Expression of Id1 in human breast cancer tumors and lung metastases. (A) Representative Id1 immunohistochemistry images of non-TN and TN primary breast tumors, acquired with an initial magnification of ×20. (B) Id1 immunohistochemistry of a matched primary tumor and lung metastasis pair. Nine of these pairs were analyzed, as discussed in the text. (Original magnification: ×20.) (C) Compiled results of Id1 immunohistochemistry performed on 133 primary breast tumors. Frequencies of Id1 positivity in endothelial cells and in cancer cells are shown. (D) Tabulated results of Id1 immunohistochemistry of 39 breast cancer lung metastases, with subset analysis based on hormone receptor expression. P values were calculated by a two-tailed Fisher's exact test.
Fig. 2.
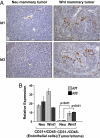
Id1 and Id3 expression in transgenic mouse mammary tumor models. (A) Representative Id1 and Id3 immunohistochemistry images of mammary tumors from MMTV-Neu and MMTV-Wnt1 mice. (Original magnification: ×20.) (B) Primary Neu and Wnt1 tumors were dissociated into single cell suspensions and separated by FACS by using the CD31 and CD45 cell surface markers. The CD31+/CD45− fraction is enriched for endothelial cells, and the CD31−/CD45− fraction is composed mostly of tumor cells but may also contain a minority of stromal cells. Total RNA was extracted from these samples, and Id1 and Id3 mRNA levels normalized to GAPDH were measured by qRT-PCR. Normalized expression values are shown relative to those obtained in the Neu tumor CD31−/CD45− fraction. Samples were analyzed in quadruplicate, and error bars represent 95% confidence intervals. P values are based on a two-tailed Student's t test.
Fig. 3.
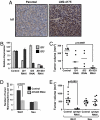
Functional requirement for Id1/Id3 in tumor-initiating capacity. (A) Id1 immunohistochemistry of unselected MDA-MB-231 parental cells and highly lung metastatic LM2 cells grown orthotopically in the mammary fat pad of an immunocompromised mouse. (Original magnification: ×20.) (B) qRT-PCR analysis of Id1 and Id3 expression in LM2 cells retrovirally transduced with short hairpin constructs targeting Id1, Id3, or both genes. Error bars indicate 95% confidence intervals of samples assayed in quadruplicate. (C) One thousand control or ID knockdown LM2 cells were injected orthotopically into the mammary glands of immunocompromised mice. Tumor volumes were assessed at 9 weeks after inoculation. The P value is based on a two-tailed t test. (D) Mammosphere-initiating capacity of primary Wnt1 and Neu tumor cells transduced and sorted for shId1/Id3 or shLuc expression. Cells were incubated in suspension at a concentration of 1,000 cells per ml, and mammospheres were counted after 14 days of culture. Error bars represent the standard error of the mean. The P value is based on a two-tailed t test. (E) Twenty-five thousand primary Wnt and 100,000 primary Neu tumor cells expressing short hairpins targeting luciferase or Id1/Id3 were injected into the mammary glands of immunocompromised mice. Tumor volumes at 3 months are represented for all cohorts. The P value is based on a two-tailed t test comparison.
Fig. 4.
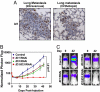
Id1/Id3 are required for lung metastasis. (A) Id1 immunohistochemistry of lung metastases generated by injection of LM2 cells intravenously and of lung metastases emerging from orthotopically implanted mammary fat pad LM2 tumors. (Original magnification: ×20.) (B) Ten thousand control LM2, Id single knockdown LM2, and combination Id1/Id3 knockdown LM2 cells were injected intravenously, and lung metastatic outgrowth was monitored by using bioluminescence imaging. The P value is based on a two-tailed rank-sum test, compared with the control cohort. (C) Bioluminescence images of representative mice from each cohort at day 0 and day 42.
Fig. 5.
Id1/Id3 mediate sustained tumor proliferation during lung colonization. (A) Reconstructed confocal images of 50-μm-thick lung sections collected 3 days after i.v. inoculation of mice with LM2–4175 cells carrying a control shRNA vector or LM2 cells with combination ID1/ID3 knockdown. Immunofluorescence staining for CD31 (red) and tumor-specific vimentin (green) show that tumor cells in both cases were able to successfully extravasate into the lung parenchyma. (Original magnification: ×63.) (B) Quantification of in vitro transendothelial migration of parental MDA-MB-231, control LM2 cells, ID1/ID3 knockdown LM2, and EREG/COX2/MMP1/2 quadruple knockdown LM2 cells (4sh-RNAi). The P values are based on a two-tailed student's t test. (C) Coimmunofluorescence for Id1 (green), PCNA (red), and nuclei (DAPI, blue) in an early stage lung metastasis generated by control LM2 cells. Blue arrowheads in the merged image indicate nuclei of cells that also stained positive for tumor-specific vimentin (not shown). Yellow arrowheads indicate tumor cells that show colocalization of Id1 and PCNA. (Original magnification: ×63.) Tabulation of 269 tumor cell nuclei that stain for PCNA and/or Id1 based on analysis of multiple independent immunofluorescence images is also shown. The P value was calculated by using a Fisher's exact test for a 2 × 2 contingency table. (D) Representative immunofluorescence images for tumor-specific vimentin (red), the cell proliferation marker Ki-67 (green), and nuclear DAPI (blue) in the lungs of mice that were intravenously inoculated with either control or ID1/ID3 combination knockdown LM2 cells. Quantification of multiple lung sections at day 5 and day 10 demonstrates significant differences in the rate of tumor cell proliferation. The P values are based on a two-tailed Student's t test. (E) A schematic representation of Id1/Id3 as mediators of proliferative functions that enable efficient secondary tumor reinitiation during lung metastasis.
Similar articles
- Targeting Id1 and Id3 by a specific peptide aptamer induces E-box promoter activity, cell cycle arrest, and apoptosis in breast cancer cells.
Mern DS, Hoppe-Seyler K, Hoppe-Seyler F, Hasskarl J, Burwinkel B. Mern DS, et al. Breast Cancer Res Treat. 2010 Dec;124(3):623-33. doi: 10.1007/s10549-010-0810-6. Epub 2010 Feb 27. Breast Cancer Res Treat. 2010. PMID: 20191379 - Targeting Id1 and Id3 inhibits peritoneal metastasis of gastric cancer.
Tsuchiya T, Okaji Y, Tsuno NH, Sakurai D, Tsuchiya N, Kawai K, Yazawa K, Asakage M, Yamada J, Yoneyama S, Kitayama J, Osada T, Watanabe T, Tokunaga K, Takahashi K, Nagawa H. Tsuchiya T, et al. Cancer Sci. 2005 Nov;96(11):784-90. doi: 10.1111/j.1349-7006.2005.00113.x. Cancer Sci. 2005. PMID: 16271072 Free PMC article. - Id1 and Id3 co-expression correlates with clinical outcome in stage III-N2 non-small cell lung cancer patients treated with definitive chemoradiotherapy.
Castañon E, Bosch-Barrera J, López I, Collado V, Moreno M, López-Picazo JM, Arbea L, Lozano MD, Calvo A, Gil-Bazo I. Castañon E, et al. J Transl Med. 2013 Jan 11;11:13. doi: 10.1186/1479-5876-11-13. J Transl Med. 2013. PMID: 23311395 Free PMC article. - Inhibitor of differentiation 1 (Id1) and Id3 proteins play different roles in TGFβ effects on cell proliferation and migration in prostate cancer cells.
Strong N, Millena AC, Walker L, Chaudhary J, Khan SA. Strong N, et al. Prostate. 2013 May;73(6):624-33. doi: 10.1002/pros.22603. Epub 2012 Oct 11. Prostate. 2013. PMID: 23060149 Free PMC article. - Inhibitor of differentiation 4 (ID4): From development to cancer.
Patel D, Morton DJ, Carey J, Havrda MC, Chaudhary J. Patel D, et al. Biochim Biophys Acta. 2015 Jan;1855(1):92-103. doi: 10.1016/j.bbcan.2014.12.002. Epub 2014 Dec 12. Biochim Biophys Acta. 2015. PMID: 25512197 Free PMC article. Review.
Cited by
- A liquid biopsy assay for the noninvasive detection of lymph node metastases in T1 lung adenocarcinoma.
Li X, Gu Y, Hu B, Shao MM, Li H. Li X, et al. Thorac Cancer. 2024 Jun;15(16):1312-1319. doi: 10.1111/1759-7714.15315. Epub 2024 Apr 29. Thorac Cancer. 2024. PMID: 38682829 Free PMC article. - Functional Enrichment Analysis of Tumor Microenvironment-Driven Molecular Alterations That Facilitate Epithelial-to-Mesenchymal Transition and Distant Metastasis.
Abdolahi M, Ghaedi Talkhounche P, Derakhshan Nazari MH, Hosseininia HS, Khoshdel-Rad N, Ebrahimi Sadrabadi A. Abdolahi M, et al. Bioinform Biol Insights. 2024 Feb 4;18:11779322241227722. doi: 10.1177/11779322241227722. eCollection 2024. Bioinform Biol Insights. 2024. PMID: 38318286 Free PMC article. - ID2 promotes tumor progression and metastasis in thyroid cancer.
Deng Z, Xu M, Ding Z, Kong J, Liu J, Zhang Z, Cao P. Deng Z, et al. Endocrine. 2024 Jun;84(3):1051-1063. doi: 10.1007/s12020-023-03674-3. Epub 2024 Jan 10. Endocrine. 2024. PMID: 38195969 Free PMC article. - Proactive and reactive roles of TGF-β in cancer.
Kuburich NA, Sabapathy T, Demestichas BR, Maddela JJ, den Hollander P, Mani SA. Kuburich NA, et al. Semin Cancer Biol. 2023 Oct;95:120-139. doi: 10.1016/j.semcancer.2023.08.002. Epub 2023 Aug 11. Semin Cancer Biol. 2023. PMID: 37572731 Free PMC article. Review. - Promyelocytic leukemia protein regulates angiogenesis and epithelial-mesenchymal transition to limit metastasis in MDA-MB-231 breast cancer cells.
Vogiatzoglou AP, Spanou S, Sachini N, Drakos E, Nikolaou C, Makatounakis T, Kretsovali A, Papamatheakis J. Vogiatzoglou AP, et al. Mol Oncol. 2023 Oct;17(10):2090-2108. doi: 10.1002/1878-0261.13501. Epub 2023 Sep 4. Mol Oncol. 2023. PMID: 37518985 Free PMC article.
References
- Fidler IJ. Nat Rev Cancer. 2003;3:453–458. - PubMed
- Gupta GP, Massague J. Cell. 2006;127:679–695. - PubMed
- Pardal R, Clarke MF, Morrison SJ. Nat Rev Cancer. 2003;3:895–902. - PubMed
- Bonnet D, Dick JE. Nat Med. 1997;3:730–737. - PubMed
- Radtke F, Clevers H. Science. 2005;307:1904–1909. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM07739/GM/NIGMS NIH HHS/United States
- P01-94060/PHS HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- P01 CA094060/CA/NCI NIH HHS/United States
- T32 GM007739/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous