MicroRNA targets in immune genes and the Dicer/Argonaute and ARE machinery components - PubMed (original) (raw)
MicroRNA targets in immune genes and the Dicer/Argonaute and ARE machinery components
Ananthi J Asirvatham et al. Mol Immunol. 2008 Apr.
Abstract
We studied 613 genes which regulate immunity and, utilizing predictive algorithms, identified 285 genes as microRNA (miRNA or miR) targets. Of these, approximately 250 are newly predicted gene-miR interactions. The frequency of predicted miRNA binding sites in immune gene 3'UTRs indicated preferential targeting of immune genes compared to the genome. Major targets include transcription factors, cofactors and chromatin modifiers whereas upstream factors, such as ligands and receptors (cytokines, chemokines and TLRs), were, in general, non-targets. About 10% of the immune genes were 'hubs' with eight or more different miRNAs predicted to target their 3'UTRs. Hubs were focused on certain key immune genes, such as BCL6, SMAD7, BLIMP1, NFAT5, EP300 and others. NF-kappaB and p53 do not themselves have binding sites for miRNAs but rather these pathways are targeted by miRNAs at downstream sites. MHC class II genes lacked miRNA targets but binding sites were identified in the CIITA gene and were shown experimentally to repress IFN-gamma-induced MHC class II activation. Unexpectedly, factors involved in regulating message stability via AU-rich elements (ARE) were heavily targeted. Moreover, multiple components involved in the generation and effector functions of miRNAs (Dicer and Argonautes) were themselves miRNA targets suggesting that a subset of miRNAs may indirectly control their own production as well as other miRNAs.
Figures
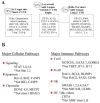
Figure 1. Examples of miRNA targeting of immune genes
A, Genes were placed in three categories according to the number of miRNA binding sites in the 3′UTR. B, MiRNA regulation of major cellular and immune pathways showing the contrast between genes which are targets versus non-targets in the same general pathway. (* indicates genes in the pathway that do not have miRNA target sites)
Figure 2. Cellular distribution of miRNA targets in immune pathways
The protein targets are classified based on their cellular localization in four categories. The percentages are based on the number of genes in each location predicted to be targeted by miRNAs by at least 2 algorithms. A number of important pathways (RNA stability, apoptosis and RNAi pathway) could not be distributed into these spatial categories and were not included in the analysis. Figure patterned after the neuronal pathways reported by Cui, Q. et al., 2006.
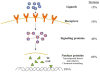
Figure 3. MiRNAs and MAP Kinase may indirectly regulate multiple inflammatory genes by regulating the components of the ARE machinery
The expression of multiple inflammatory molecules and immune genes is known to be regulated by mRNA stability mediated by AU-rich elements located in their 3′UTRs. The predictions suggest that the expression of the machinery components for ARE are regulated by miRNA. The specific miRNAs targeting the ARE machinery are listed in Supplemental Table 7.
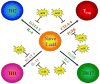
Figure 4. Potential miRNA regulation of T cell differentiation
The schematic diagram describes the primary effectors of Th1, Th2, Th17 and Treg differentiation pathways and indicates the predicted miRNA targeting of these pathways. The specific miRNAs predicted to target each of the effector genes can be found in Supplemental Table 3.
Figure 5. Role of miRNA in chromatin regulation
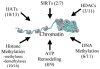
Multiple pathways contribute to the regulation of chromatin structure and thereby gene expression. For each category of chromatin regulator, the number of genes found to have miRNA targets (consensus of two algorithms) and the total number of genes analyzed in that category are shown in parentheses. Our predictions suggest significant contributions to chromatin regulation in multiple pathways from miRNA. See Supplemental Table 11 for specific miRNA targets.
Figure 6. MiRNA regulation of CIITA and MHC class II induction
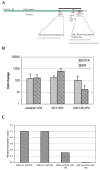
A, Schematic representation of the CIITA transcript illustrating the predicted miRNA binding sites and AREs in the 3′UTR. The black bar indicates the fragment present in the pMir-Luc-CIITA reporter construct. B, miR-145 inhibits the IFN- γ induction of HLA-DR in HeLa cells. HeLa cultures were transfected with pre-miR-145, treated with IFN- γ and, after 24hrs, total RNA (mirVana, Ambion) was analyzed for CIITA and HLA-DR α expression by real time RT-PCR. The negative control, NC1, non-targeting pre-miR, and untransfected cells are shown as controls. C, miR-145 and 198 inhibit a heterologous reporter when the CIITA 3′UTR is present. HeLa cells were transfected with reporter constructs carrying 1kb of the CIITA 3′UTR (pMir-Luc-CIITA) or no insert (pMir-Luc) along with 100nM pre-miR-145 or 198 and, 24hr later, whole cell lysates were analyzed for luciferase activity. Co-transfection with pMIR- βgal allowed normalization to β-galactosidase activity.
Similar articles
- Histone deacetylase 1/mSin3A disrupts gamma interferon-induced CIITA function and major histocompatibility complex class II enhanceosome formation.
Zika E, Greer SF, Zhu XS, Ting JP. Zika E, et al. Mol Cell Biol. 2003 May;23(9):3091-102. doi: 10.1128/MCB.23.9.3091-3102.2003. Mol Cell Biol. 2003. PMID: 12697811 Free PMC article. - Expression of the MHC class II transactivator (CIITA) type IV promoter in B lymphocytes and regulation by IFN-gamma.
Piskurich JF, Gilbert CA, Ashley BD, Zhao M, Chen H, Wu J, Bolick SC, Wright KL. Piskurich JF, et al. Mol Immunol. 2006 Feb;43(6):519-28. doi: 10.1016/j.molimm.2005.05.005. Epub 2005 Jun 13. Mol Immunol. 2006. PMID: 15950283 Free PMC article. - Regulation of the MIR155 host gene in physiological and pathological processes.
Elton TS, Selemon H, Elton SM, Parinandi NL. Elton TS, et al. Gene. 2013 Dec 10;532(1):1-12. doi: 10.1016/j.gene.2012.12.009. Epub 2012 Dec 14. Gene. 2013. PMID: 23246696 Review. - Expression of MHC II genes.
Drozina G, Kohoutek J, Jabrane-Ferrat N, Peterlin BM. Drozina G, et al. Curr Top Microbiol Immunol. 2005;290:147-70. doi: 10.1007/3-540-26363-2_7. Curr Top Microbiol Immunol. 2005. PMID: 16480042 Review.
Cited by
- MicroRNAs in blood and cerebrospinal fluid as diagnostic biomarkers of multiple sclerosis and to monitor disease progression.
Martinez B, Peplow PV. Martinez B, et al. Neural Regen Res. 2020 Apr;15(4):606-619. doi: 10.4103/1673-5374.266905. Neural Regen Res. 2020. PMID: 31638082 Free PMC article. Review. - Tie2cre-induced inactivation of the miRNA-processing enzyme Dicer disrupts invariant NKT cell development.
Zhou L, Seo KH, He HZ, Pacholczyk R, Meng DM, Li CG, Xu J, She JX, Dong Z, Mi QS. Zhou L, et al. Proc Natl Acad Sci U S A. 2009 Jun 23;106(25):10266-71. doi: 10.1073/pnas.0811119106. Epub 2009 Jun 9. Proc Natl Acad Sci U S A. 2009. PMID: 19509335 Free PMC article. - MicroRNAs: control and loss of control in human physiology and disease.
Li M, Marin-Muller C, Bharadwaj U, Chow KH, Yao Q, Chen C. Li M, et al. World J Surg. 2009 Apr;33(4):667-84. doi: 10.1007/s00268-008-9836-x. World J Surg. 2009. PMID: 19030926 Free PMC article. Review. - Helicobacter pylori Dampens HLA-II Expression on Macrophages via the Up-Regulation of miRNAs Targeting CIITA.
Codolo G, Toffoletto M, Chemello F, Coletta S, Soler Teixidor G, Battaggia G, Munari G, Fassan M, Cagnin S, de Bernard M. Codolo G, et al. Front Immunol. 2020 Jan 8;10:2923. doi: 10.3389/fimmu.2019.02923. eCollection 2019. Front Immunol. 2020. PMID: 31969878 Free PMC article. - MicroRNAs: the fine-tuners of Toll-like receptor signalling.
O'Neill LA, Sheedy FJ, McCoy CE. O'Neill LA, et al. Nat Rev Immunol. 2011 Mar;11(3):163-75. doi: 10.1038/nri2957. Epub 2011 Feb 18. Nat Rev Immunol. 2011. PMID: 21331081 Review.
References
- Anastasiou D, Krek W. SIRT1: Linking Adaptive Cellular Responses to Aging-Associated Changes in Organismal Physiology. Physiology. 2006;21:404–410. - PubMed
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
- Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2:S4–11. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA016056/CA/NCI NIH HHS/United States
- CA16056/CA/NCI NIH HHS/United States
- R01 HD017013-19/HD/NICHD NIH HHS/United States
- R01 HD017013/HD/NICHD NIH HHS/United States
- HD17013/HD/NICHD NIH HHS/United States
- R01 CA124971/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous