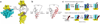tRNA-mRNA mimicry drives translation initiation from a viral IRES - PubMed (original) (raw)
tRNA-mRNA mimicry drives translation initiation from a viral IRES
David A Costantino et al. Nat Struct Mol Biol. 2008 Jan.
Abstract
Internal ribosome entry site (IRES) RNAs initiate protein synthesis in eukaryotic cells by a noncanonical cap-independent mechanism. IRESes are critical for many pathogenic viruses, but efforts to understand their function are complicated by the diversity of IRES sequences as well as by limited high-resolution structural information. The intergenic region (IGR) IRESes of the Dicistroviridae viruses are powerful model systems to begin to understand IRES function. Here we present the crystal structure of a Dicistroviridae IGR IRES domain that interacts with the ribosome's decoding groove. We find that this RNA domain precisely mimics the transfer RNA anticodon-messenger RNA codon interaction, and its modeled orientation on the ribosome helps explain translocation without peptide bond formation. When combined with a previous structure, this work completes the first high-resolution description of an IRES RNA and provides insight into how RNAs can manipulate complex biological machines.
Figures
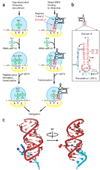
Figure 1. The P site domain of the Dicistroviridae intergenic region IRESes
(a) Comparison of canonical initiation of protein synthesis compared with that driven by the Dicistroviridae IGR IRES RNA. The steps of initial ribosome recruitment and placement on the message are not shown. 40S subunit, yellow, 60S subunit, blue; tRNA, green; ribosome binding domain (regions 1 + 2) of the IRES, black; domain 3 of the IRES, red. Locations of the A, P and E sites are labeled. (b) Secondary structure of domain 3 from the CrPV IGR IRES. The orientation of this domain has been flipped relative to panel a to reflect how it generally is depicted in the literature. Nucleotides in lowercase are not included in our crystal construct. Gray-shaded boxes, nucleotides that are conserved in six or more of seven members of this IRES class (Supplementary Fig. 1). Dashed boxes, mutations that introduced a heavy atom binding site for obtaining crystallographic phase information (Supplementary Fig. 2). (c) Two views (90° rotation) of the crystal structure of domain 3 of the CrPV IGR IRES, colored to match the secondary structure of panel b. Nts 6203–6209 are not shown because their structure was affected by crystal packing (Supplementary Fig. 3).
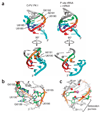
Figure 2. Structural details and comparison to authentic tRNA-mRNA interactions
(a) Left, two views of PK I of domain 3 that mimics the anticodon-codon interaction; right, corresponding views of an authentic initiator tRNA anticodon–mRNA codon interaction in the P site of a ribosome (from PDB 2J00). Corresponding bases in the two structures are colored to match each other. Cyan in tRNA–mRNA structure (right), mRNA; cyan in domain 3 structure (left), 3′ end of the IRES RNA that mimics mRNA. (b) Interactions that stabilize the structure, with highly conserved nucleotides (Fig. 1b,c) colored, more variable nucleotides gray, and the U6186 cross-loop hydrogen bond indicated with an asterisk. (c) Location of a constrained iridium(III) hexammine cation (magenta) in the major groove of the anticodon loop of the P site domain structure. Highly conserved bases are colored and variable bases are gray. Partially negatively charged functional groups, including base carbonyl and phosphate oxygens, are in red. Dashed line, location of the U6186 cross-loop hydrogen bond to G6189’s phosphate. The three ‘anticodon’ bases that are conserved as purines are indicated.
Figure 3. Translation initiation assays for mutant CrPV IGR IRES RNAs
(a) Diagram of a dual luciferase construct, with the firefly (F) and renilla (R) luciferase genes shown. (b–e) Bar graphs showing the average F/R ratios of mutant CrPV IGR IRES RNAs compared with wild type and a negative control IRES (ΔPK I/ΔPK III). ‘Triple’ refers to the mutant containing U6185C + U6186A + G6192C. Error bars, s.e.m. from at least four experiments. Values are given in Table 1.
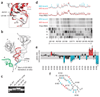
Figure 4. Model and probing of domain 3 in the ribosome P site
(a) The part of CrPV IGR IRES domain 3 that mimics the tRNA anticodon loop (red) aligned with the structure of the anticodon loop of an authentic P site–bound tRNAMet (gray, from PDB 2J00). Dashed box, portion of the structure that is a precise mimic of the initiator tRNA anticodon loop. (b) View of the structure of CrPV domain 3 docked into its predicted position within the decoding groove, aligned with P site tRNA as in panel a. The CrPV domain 3 is in red and cyan, a portion of A site tRNA is in green, and P and E site tRNAs and mRNA are in gray. During CrPV IGR IRES-driven initiation, only the A site tRNA is present. (c) Native gel of mutants d3_6190–6191_comp (U6190A + A6191U) and d3_Δ6191 next to wild-type domain 3 in the presence of 10 mM MgCl2. (d) A portion of an example gel of PSIV IGR IRES domain 3 modified with NMIA. Lanes containing free IRES RNA, 40S-bound IRES RNA and 80S-bound IRES RNA are shown over a dideoxy sequencing ladder. For clarity, this figure was constructed by splicing lanes from a single gel. Above the gel is a trace quantification of each lane. Some of the nucleotides with strong changes are labeled. (e) Graph of multiple SHAPE probing experiments quantified and analyzed. Gray dashed box, one s.d. between multiple measurements. The difference in modification between 40S-bound and 80S-bound IRES is shown, and changes greater than one s.d. are colored; red bars above the zero line, nucleotides that are modified more in the context of the 80S ribosome; blue bars under the line, those that are modified less. (f) Changes of panel e overlaid on the PSIV IGR IRES domain 3 secondary structure.
Figure 5. Docking of both IRES domain structures into a cryo-EM reconstruction and hypothesized mimicry mechanism
(a) Three views of the crystal structure of the ribosome binding domain of the PSIV IRES and the domain 3 structure docked into the cryo-EM structure of the CrPV IRES on a human 80S ribosome. At left is a view from the ‘top’ of the 80S; at top right the 60S subunit has been removed computationally, and at bottom right is this same view rotated 90° with the 60S subunit included. IGR IRES contacts to P and E site interaction sites are indicated. (b) Comparison of the angle between docked domain 3 and P site tRNA (from PDB 2J00), and P/E hybrid state and P site tRNAs (from PDB 1Z03). (c) Proposed mechanistic model of tRNA hybrid-state mimicry by the Dicistroviridae IRES. Top, canonical method of translocation in which the tRNAs adopt a hybrid state after peptide bond formation but before translocation. Amino acids in the peptide chain, gray circles, P and A site tRNAs, red and green outlines, respectively. 40S subunit, yellow; 60S, blue. Darker shades of blue, conformational changes in the subunit that are associated with the tRNA hybrid states. Bottom, proposed corresponding steps in the IGR IRES mechanism that mimic the hybrid state. IRES, red; the two IRES domains are indicated. Figure is based on figure from ref. .
Similar articles
- Modular domains of the Dicistroviridae intergenic internal ribosome entry site.
Jang CJ, Jan E. Jang CJ, et al. RNA. 2010 Jun;16(6):1182-95. doi: 10.1261/rna.2044610. Epub 2010 Apr 27. RNA. 2010. PMID: 20423979 Free PMC article. - Taura syndrome virus IRES initiates translation by binding its tRNA-mRNA-like structural element in the ribosomal decoding center.
Koh CS, Brilot AF, Grigorieff N, Korostelev AA. Koh CS, et al. Proc Natl Acad Sci U S A. 2014 Jun 24;111(25):9139-44. doi: 10.1073/pnas.1406335111. Epub 2014 Jun 9. Proc Natl Acad Sci U S A. 2014. PMID: 24927574 Free PMC article. - Dual tRNA mimicry in the Cricket Paralysis Virus IRES uncovers an unexpected similarity with the Hepatitis C Virus IRES.
Pisareva VP, Pisarev AV, Fernández IS. Pisareva VP, et al. Elife. 2018 Jun 1;7:e34062. doi: 10.7554/eLife.34062. Elife. 2018. PMID: 29856316 Free PMC article. - RNA structure-based ribosome recruitment: lessons from the Dicistroviridae intergenic region IRESes.
Pfingsten JS, Kieft JS. Pfingsten JS, et al. RNA. 2008 Jul;14(7):1255-63. doi: 10.1261/rna.987808. Epub 2008 May 30. RNA. 2008. PMID: 18515544 Free PMC article. Review. - tRNA-mimicry in IRES-mediated translation and recoding.
Butcher SE, Jan E. Butcher SE, et al. RNA Biol. 2016 Nov;13(11):1068-1074. doi: 10.1080/15476286.2016.1219833. Epub 2016 Aug 11. RNA Biol. 2016. PMID: 27654067 Free PMC article. Review.
Cited by
- Cryo-EM of ribosomal 80S complexes with termination factors reveals the translocated cricket paralysis virus IRES.
Muhs M, Hilal T, Mielke T, Skabkin MA, Sanbonmatsu KY, Pestova TV, Spahn CM. Muhs M, et al. Mol Cell. 2015 Feb 5;57(3):422-32. doi: 10.1016/j.molcel.2014.12.016. Epub 2015 Jan 15. Mol Cell. 2015. PMID: 25601755 Free PMC article. - Virus versus host cell translation love and hate stories.
Komarova AV, Haenni AL, Ramírez BC. Komarova AV, et al. Adv Virus Res. 2009;73:99-170. doi: 10.1016/S0065-3527(09)73003-9. Adv Virus Res. 2009. PMID: 19695382 Free PMC article. Review. - Structural basis for the biological relevance of the invariant apical stem in IRES-mediated translation.
Fernández N, Fernandez-Miragall O, Ramajo J, García-Sacristán A, Bellora N, Eyras E, Briones C, Martínez-Salas E. Fernández N, et al. Nucleic Acids Res. 2011 Oct;39(19):8572-85. doi: 10.1093/nar/gkr560. Epub 2011 Jul 8. Nucleic Acids Res. 2011. PMID: 21742761 Free PMC article. - Structural determinants of an internal ribosome entry site that direct translational reading frame selection.
Ren Q, Au HH, Wang QS, Lee S, Jan E. Ren Q, et al. Nucleic Acids Res. 2014 Aug;42(14):9366-82. doi: 10.1093/nar/gku622. Epub 2014 Jul 18. Nucleic Acids Res. 2014. PMID: 25038250 Free PMC article. - Dynamics of IRES-mediated translation.
Johnson AG, Grosely R, Petrov AN, Puglisi JD. Johnson AG, et al. Philos Trans R Soc Lond B Biol Sci. 2017 Mar 19;372(1716):20160177. doi: 10.1098/rstb.2016.0177. Philos Trans R Soc Lond B Biol Sci. 2017. PMID: 28138065 Free PMC article. Review.
References
- Hershey JWB, Merrick WC. Pathway and mechanism of initiation of protein synthesis. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor, New York, USA: Cold Spring Harbor Laboratory Press; 2000. pp. 33–88.
- Stoneley M, Willis AE. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. - PubMed
- Jackson RJ. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem. Soc. Trans. 2005;33:1231–1241. - PubMed
- Schuler M, et al. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat. Struct. Mol. Biol. 2006;13:1092–1096. - PubMed
- Spahn CM, et al. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes; the IRES functions as an RNA-based translation factor. Cell. 2004;118:465–475. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM072560/GM/NIGMS NIH HHS/United States
- R01 GM072560-03/GM/NIGMS NIH HHS/United States
- R01 GM072560-01/GM/NIGMS NIH HHS/United States
- R03 AI072187/AI/NIAID NIH HHS/United States
- R01 GM072560-04/GM/NIGMS NIH HHS/United States
- AI072187/AI/NIAID NIH HHS/United States
- R03 AI072187-01A1/AI/NIAID NIH HHS/United States
- R03 AI072187-02/AI/NIAID NIH HHS/United States
- R01 GM072560-05/GM/NIGMS NIH HHS/United States
- GM072560/GM/NIGMS NIH HHS/United States
- R01 GM072560-02/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous