Lysogenic transfer of group A Streptococcus superantigen gene among Streptococci - PubMed (original) (raw)
Lysogenic transfer of group A Streptococcus superantigen gene among Streptococci
Ivo Vojtek et al. J Infect Dis. 2008.
Abstract
A group A Streptococcus (GAS) isolate, serotype M12, recovered from a patient with streptococcal toxic shock syndrome was analyzed for superantigen-carrying prophages, revealing phi149, which encodes superantigen SSA. Sequence analysis of the att-L proximal region of phi149 showed that the phage had a mosaic nature. Remarkably, we successfully obtained lysogenic conversion of GAS clinical isolates of various M serotypes (M1, M3, M5, M12, M19, M28, and M94), as well as of group C Streptococcus equisimilis (GCSE) clinical isolates, via transfer of a recombinant phage phi149::Km(r). Phage phi149::Km(r) from selected lysogenized GAS and GCSE strains could be transferred back to M12 GAS strains. Our data indicate that horizontal transfer of lysogenic phages among GAS can occur across the M-type barrier; these data also provide further support for the hypothesis that toxigenic conversion can occur via lysogeny between species. Streptococci might employ this mechanism specifically to allow more efficient adaptation to changing host challenges, potentially leading to fitter and more virulent clones.
Figures
Figure 1
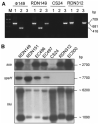
Gel electrophoresis patterns demonstrating ex vivo lysogenic transfer of _ssa-3_– carrying phage  149 to group A Streptococcus (GAS) clinical isolates of type M12. A, Results of polymerase chain reaction analysis of isolated phage DNA from strain RDN149 and genomic DNA from phage donor strain RDN149, phage indicator strain CS24, and lysogenized strain RDN312 with primers specific to the ssa gene (lane 1), the speH gene (lane 2) and the hki gene (lane 3). hki encoding a putative histidine kinase served as positive control for GAS genomic DNA. M, 1-kb DNA ladder (Fermentas Life Sciences). The 709-, 681-, and 416-bp DNA fragments correspond to the _ssa_-, _hki_-, and _speH_-specific amplified products, respectively. B, Results of Southern blot analysis of _Hin_dIII-digested genomic DNA of phage donor strain RDN149; phage indicator strains RDN151 and CS24; and lysogenized strains EC496, EC497, RDN312, and EC500, with _α_-32P labeled DNA probes specific to the ssa, speH, and hki genes. The approximate sizes of the hybridizing fragments are indicated in bp.
149 to group A Streptococcus (GAS) clinical isolates of type M12. A, Results of polymerase chain reaction analysis of isolated phage DNA from strain RDN149 and genomic DNA from phage donor strain RDN149, phage indicator strain CS24, and lysogenized strain RDN312 with primers specific to the ssa gene (lane 1), the speH gene (lane 2) and the hki gene (lane 3). hki encoding a putative histidine kinase served as positive control for GAS genomic DNA. M, 1-kb DNA ladder (Fermentas Life Sciences). The 709-, 681-, and 416-bp DNA fragments correspond to the _ssa_-, _hki_-, and _speH_-specific amplified products, respectively. B, Results of Southern blot analysis of _Hin_dIII-digested genomic DNA of phage donor strain RDN149; phage indicator strains RDN151 and CS24; and lysogenized strains EC496, EC497, RDN312, and EC500, with _α_-32P labeled DNA probes specific to the ssa, speH, and hki genes. The approximate sizes of the hybridizing fragments are indicated in bp.
Figure 2
Comparison of the att-L proximal region of _ssa-3_–carrying phage  149 with the att-L proximal regions of ssa-1 –carrying phages
149 with the att-L proximal regions of ssa-1 –carrying phages  315.2 (MGAS315; M3) and SPsP6 (SSI-1, M3) and _mf3_-carrying phages
315.2 (MGAS315; M3) and SPsP6 (SSI-1, M3) and _mf3_-carrying phages  315.3 (MGAS315; M3), SPsP4 (SSI-1; M3),
315.3 (MGAS315; M3), SPsP4 (SSI-1; M3),  370.2 (SF370; M1), and
370.2 (SF370; M1), and  370.2-like (MGAS8232; M18). The shaded areas indicate the regions of similarity with the percentage of nucleotide identity given. The 34 bp IS_1239_-related sequence located 103 bp upstream of the ssa coding sequence is shown. Annotated open reading frames refer to the genomes of MGAS315 strain (
370.2-like (MGAS8232; M18). The shaded areas indicate the regions of similarity with the percentage of nucleotide identity given. The 34 bp IS_1239_-related sequence located 103 bp upstream of the ssa coding sequence is shown. Annotated open reading frames refer to the genomes of MGAS315 strain ( 315.2) and SF370 (
315.2) and SF370 ( 370.2). Small, black arrows, positions of primers.
370.2). Small, black arrows, positions of primers.
Figure 3
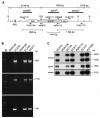
Analysis of the broad host range of recombinant phage  149::Kmr. A, Representative restriction map of the att-L proximal region of
149::Kmr. A, Representative restriction map of the att-L proximal region of  149::Kmr. Genes are drawn to scale. Small, black arrows, positions of primers. Sizes of PCR and hybridizing restriction fragments are indicated in bp. B, Results of polymerase chain reaction analysis of genomic DNA from
149::Kmr. Genes are drawn to scale. Small, black arrows, positions of primers. Sizes of PCR and hybridizing restriction fragments are indicated in bp. B, Results of polymerase chain reaction analysis of genomic DNA from  149 donor strain RDN149,
149 donor strain RDN149,  149::Kmr donor strain EC516, recipient strain RDN156 (M28), and lysogenized strains EC749 and EC917, with primers OLEC178 and oliRN70 (top), oliRN69 and OLEC179 (middle), and oliRN111 and oliRN110 (bottom). M, 1-kb DNA ladder (Fermentas Life Sciences). C, Results of Southern blot analysis of _Hin_dIII-digested genomic DNA from
149::Kmr donor strain EC516, recipient strain RDN156 (M28), and lysogenized strains EC749 and EC917, with primers OLEC178 and oliRN70 (top), oliRN69 and OLEC179 (middle), and oliRN111 and oliRN110 (bottom). M, 1-kb DNA ladder (Fermentas Life Sciences). C, Results of Southern blot analysis of _Hin_dIII-digested genomic DNA from  149::Kmr donor strain EC516, phage recipient strain RDN156 (M28) and corresponding lysogenized strains EC749 and EC917, phage recipient strain RDN116 (M94) and corresponding lysogenized strains EC727 and EC1083 with _α_-32P labelled DNA probes specific to the ssa upstream fragment (_ssa_up), aphIII gene, and ssa downstream fragment (_ssa_dw).
149::Kmr donor strain EC516, phage recipient strain RDN156 (M28) and corresponding lysogenized strains EC749 and EC917, phage recipient strain RDN116 (M94) and corresponding lysogenized strains EC727 and EC1083 with _α_-32P labelled DNA probes specific to the ssa upstream fragment (_ssa_up), aphIII gene, and ssa downstream fragment (_ssa_dw).
Similar articles
- Genomic Sequencing of High-Efficiency Transducing Streptococcal Bacteriophage A25: Consequences of Escape from Lysogeny.
McCullor K, Postoak B, Rahman M, King C, McShan WM. McCullor K, et al. J Bacteriol. 2018 Nov 6;200(23):e00358-18. doi: 10.1128/JB.00358-18. Print 2018 Dec 1. J Bacteriol. 2018. PMID: 30224437 Free PMC article. - Dissemination of the phage-associated novel superantigen gene speL in recent invasive and noninvasive Streptococcus pyogenes M3/T3 isolates in Japan.
Ikebe T, Wada A, Inagaki Y, Sugama K, Suzuki R, Tanaka D, Tamaru A, Fujinaga Y, Abe Y, Shimizu Y, Watanabe H; Working Group for Group A Streptococcus in Japan. Ikebe T, et al. Infect Immun. 2002 Jun;70(6):3227-33. doi: 10.1128/IAI.70.6.3227-3233.2002. Infect Immun. 2002. PMID: 12011018 Free PMC article. - Transcription analysis of Streptococcus thermophilus phages in the lysogenic state.
Ventura M, Bruttin A, Canchaya C, Brüssow H. Ventura M, et al. Virology. 2002 Oct 10;302(1):21-32. doi: 10.1006/viro.2002.1571. Virology. 2002. PMID: 12429513 - The fundamental contribution of phages to GAS evolution, genome diversification and strain emergence.
Banks DJ, Beres SB, Musser JM. Banks DJ, et al. Trends Microbiol. 2002 Nov;10(11):515-21. doi: 10.1016/s0966-842x(02)02461-7. Trends Microbiol. 2002. PMID: 12419616 Review. - [Lytic phages and prophages of Streptococcus suis--a review].
Tang F, Lu C. Tang F, et al. Wei Sheng Wu Xue Bao. 2015 Apr 4;55(4):389-94. Wei Sheng Wu Xue Bao. 2015. PMID: 26211312 Review. Chinese.
Cited by
- Survival Strategies of Streptococcus pyogenes in Response to Phage Infection.
Beerens D, Franch-Arroyo S, Sullivan TJ, Goosmann C, Brinkmann V, Charpentier E. Beerens D, et al. Viruses. 2021 Apr 2;13(4):612. doi: 10.3390/v13040612. Viruses. 2021. PMID: 33918348 Free PMC article. - Genome Analysis of Streptococcus pyogenes Associated with Pharyngitis and Skin Infections.
Ibrahim J, Eisen JA, Jospin G, Coil DA, Khazen G, Tokajian S. Ibrahim J, et al. PLoS One. 2016 Dec 15;11(12):e0168177. doi: 10.1371/journal.pone.0168177. eCollection 2016. PLoS One. 2016. PMID: 27977735 Free PMC article. - Streptococcus pyogenes and re-emergence of scarlet fever as a public health problem.
Wong SS, Yuen KY. Wong SS, et al. Emerg Microbes Infect. 2012 Jul;1(7):e2. doi: 10.1038/emi.2012.9. Epub 2012 Jul 11. Emerg Microbes Infect. 2012. PMID: 26038416 Free PMC article. Review. - Natural solution to antibiotic resistance: bacteriophages 'The Living Drugs'.
Jassim SA, Limoges RG. Jassim SA, et al. World J Microbiol Biotechnol. 2014 Aug;30(8):2153-70. doi: 10.1007/s11274-014-1655-7. Epub 2014 Apr 30. World J Microbiol Biotechnol. 2014. PMID: 24781265 Free PMC article. Review. - Pathogenic characterization of a cervical lymph node derived from a patient with Kawasaki disease.
Katano H, Sato S, Sekizuka T, Kinumaki A, Fukumoto H, Sato Y, Hasegawa H, Morikawa S, Saijo M, Mizutani T, Kuroda M. Katano H, et al. Int J Clin Exp Pathol. 2012;5(8):814-23. Epub 2012 Oct 1. Int J Clin Exp Pathol. 2012. PMID: 23071864 Free PMC article.
References
- Banks DJ, Beres SB, Musser JM. The fundamental contribution of phages to GAS evolution, genome diversification and strain emergence. Trends Microbiol. 2002;10:515–21. - PubMed
- Aziz RK, Ismail SA, Park HW, Kotb M. Post-proteomic identification of a novel phage-encoded streptodornase, Sda1, in invasive M1T1 Streptococcus pyogenes. Mol Microbiol. 2004;54:184–97. - PubMed
- Kotb M, Norrby-Teglund A, McGeer A. An immunogenetic and molecular basis for differences in outcomes of invasive group A streptococcal infections. Nat Med. 2002;8:1398–404. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials