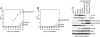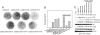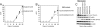Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms - PubMed (original) (raw)
Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms
Li Zhao et al. Proc Natl Acad Sci U S A. 2008.
Abstract
The phosphatidylinositol 3-kinase (PI3K) signaling pathway is up-regulated in cancer. PIK3CA, the gene coding for the catalytic subunit p110alpha of PI3K, is mutated in approximately 30% of tumors of the prostate, breast, cervix, and endometrium. The most prominent of these mutants, represented by single amino acid substitutions in the helical or kinase domain, show a gain of enzymatic function, activate AKT signaling, and induce oncogenic transformation. We have carried out a genetic and biochemical analysis of these hot-spot mutations in PIK3CA. The results of this study suggest that the helical and kinase domain mutations trigger gain of function through different mechanisms. They show different requirements for interaction with the PI3K regulatory subunit p85 and with RAS-GTP. The gain of function induced by helical domain mutations is independent of binding to p85 but requires interaction with RAS-GTP. In contrast, the kinase domain mutation is active in the absence of RAS-GTP binding but is highly dependent on the interaction with p85. We speculate that the contrasting roles of p85 and RAS-GTP in helical and kinase domain mutations reflect two distinct states of mutated p110alpha. These two states differ in mutation-induced surface charges and also may differ in conformational properties that are controlled by interactions with p85 and RAS-GTP. The two states do not appear mutually exclusive because the helical and kinase domain mutations act synergistically when present in the same p110alpha molecule. This synergism also supports the conclusion that the helical and kinase domain mutations operate by two different and independent mechanisms.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Helical domain and kinase domain mutations act synergically in cell transformation. (A and B) Focus growth curves for CEFs transfected with RCAS vectors encoding the p110α mutants. The focus growth curve of E545K/H1047R is similar to that of E542K/H1047R and is therefore not shown. EOT, number of foci per nanogram of DNA. The means for two experiments are shown. (C) Western blots comparing the protein expression levels of p110α and the phosphorylation levels of AKT and S6K. Cells were starved in basal medium, and lysates were prepared as described in Materials and Methods.
Fig. 2.
Binding to p85 is essential for H1047R-induced cell transformation. (A and B) Cell transformation (focus formation) induced by full-length or p85-binding domain deletion mutants of p110α (δp85BD-p110α). EOT by H1047R is normalized to one. (C) Western blots comparing the p110α expression levels and the phosphorylation levels of AKT and S6K. δp85BD-p110α constructs do not coimmunoprecipitate with endogenous p85α. The assays were carried out as described in Materials and Methods.
Fig. 3.
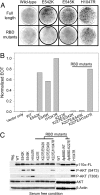
Helical domain mutations are incapable of rescuing the oncogenic activity of δp85BD-p110α H1047R. (A and B) Cell transformation induced by δp85BD-p110α constructs. EOT by δp85BD-p110α is normalized to one. (C) Western blots comparing the p110α expression levels and the phosphorylation levels of AKT and S6K. The assays were carried out as described in Materials and Methods.
Fig. 4.
Oncogenic transformation by the helical domain mutation depends on binding to RAS. (A and B) Cell transformation induced by full-length or RAS-binding mutants of p110α. EOT by H1047R is normalized to one. (C) Western blots comparing the protein expression levels of p110α and the phosphorylation levels of AKT. The assays were carried out as described in Materials and Methods.
Fig. 5.
RAS-binding mutation of p110α abolishes the activation by HRAS (G12V). CEFs were transfected with the p110α expression vector only or cotransfected with p110α and HRAS (G12V) expression vectors. Cells were maintained in a nutrient medium containing 3% FBS and 1% chicken serum and were then harvested and probed with the indicated antibodies by Western blotting.
Fig. 6.
The H1047R mutation rescues the cell-transformation phenotypes of the p110α mutants K227E/E542K and K227E/E545K from RAS-binding mutation. (A and B) Focus growth curves for CEFs transfected with RCAS vectors encoding the p110α mutants. EOT, number of foci per nanogram of DNA. The means for two experiments are shown. (C) Western blots comparing the protein expression levels of p110α and the phosphorylation levels of AKT. The assays were carried out as described in Materials and Methods.
Fig. 7.
A schematic summary of mutant properties. p85BD, p85-binding domain; RBD, Ras-binding domain; C2, C2 domain. The approximate locations of the point mutations are indicated by inverted triangles, and the deletion in the p85BD is marked by a truncated alias of the p85BD. Cell-transforming activity is qualitatively denoted by + and −, and synergistic activity is denoted by +!.
Similar articles
- Hot-spot mutations in p110alpha of phosphatidylinositol 3-kinase (pI3K): differential interactions with the regulatory subunit p85 and with RAS.
Zhao L, Vogt PK. Zhao L, et al. Cell Cycle. 2010 Feb 1;9(3):596-600. doi: 10.4161/cc.9.3.10599. Cell Cycle. 2010. PMID: 20009532 Free PMC article. - Gain of interaction with IRS1 by p110α-helical domain mutants is crucial for their oncogenic functions.
Hao Y, Wang C, Cao B, Hirsch BM, Song J, Markowitz SD, Ewing RM, Sedwick D, Liu L, Zheng W, Wang Z. Hao Y, et al. Cancer Cell. 2013 May 13;23(5):583-93. doi: 10.1016/j.ccr.2013.03.021. Epub 2013 May 2. Cancer Cell. 2013. PMID: 23643389 Free PMC article. - Human tumor mutants in the p110alpha subunit of PI3K.
Liu Z, Roberts TM. Liu Z, et al. Cell Cycle. 2006 Apr;5(7):675-7. doi: 10.4161/cc.5.7.2605. Epub 2006 Apr 1. Cell Cycle. 2006. PMID: 16627990 Review. - Differential enhancement of breast cancer cell motility and metastasis by helical and kinase domain mutations of class IA phosphoinositide 3-kinase.
Pang H, Flinn R, Patsialou A, Wyckoff J, Roussos ET, Wu H, Pozzuto M, Goswami S, Condeelis JS, Bresnick AR, Segall JE, Backer JM. Pang H, et al. Cancer Res. 2009 Dec 1;69(23):8868-76. doi: 10.1158/0008-5472.CAN-09-1968. Epub 2009 Nov 10. Cancer Res. 2009. PMID: 19903845 Free PMC article. - Targeting the protein-protein interaction between IRS1 and mutant p110α for cancer therapy.
Hao Y, Zhao S, Wang Z. Hao Y, et al. Toxicol Pathol. 2014 Jan;42(1):140-7. doi: 10.1177/0192623313506794. Epub 2013 Oct 31. Toxicol Pathol. 2014. PMID: 24178578 Free PMC article. Review.
Cited by
- Somatic profiling of the epidermal growth factor receptor pathway in tumors from patients with advanced colorectal cancer treated with chemotherapy ± cetuximab.
Smith CG, Fisher D, Claes B, Maughan TS, Idziaszczyk S, Peuteman G, Harris R, James MD, Meade A, Jasani B, Adams RA, Kenny S, Kaplan R, Lambrechts D, Cheadle JP. Smith CG, et al. Clin Cancer Res. 2013 Aug 1;19(15):4104-13. doi: 10.1158/1078-0432.CCR-12-2581. Epub 2013 Jun 5. Clin Cancer Res. 2013. PMID: 23741067 Free PMC article. - Defining biomarkers to predict sensitivity to PI3K/Akt/mTOR pathway inhibitors in breast cancer.
Gonzalez-Angulo AM, Blumenschein GR Jr. Gonzalez-Angulo AM, et al. Cancer Treat Rev. 2013 Jun;39(4):313-20. doi: 10.1016/j.ctrv.2012.11.002. Epub 2012 Dec 6. Cancer Treat Rev. 2013. PMID: 23218708 Free PMC article. - Allosteric activation of PI3Kα by oncogenic mutations.
Burke JE, Perisic O, Williams RL. Burke JE, et al. Oncotarget. 2013 Feb;4(2):180-1. doi: 10.18632/oncotarget.913. Oncotarget. 2013. PMID: 23563630 Free PMC article. No abstract available. - Optimal targeting of HER2-PI3K signaling in breast cancer: mechanistic insights and clinical implications.
Rexer BN, Arteaga CL. Rexer BN, et al. Cancer Res. 2013 Jul 1;73(13):3817-20. doi: 10.1158/0008-5472.CAN-13-0687. Epub 2013 Jun 21. Cancer Res. 2013. PMID: 23794708 Free PMC article. Review. - mTORC1 inhibition is required for sensitivity to PI3K p110α inhibitors in PIK3CA-mutant breast cancer.
Elkabets M, Vora S, Juric D, Morse N, Mino-Kenudson M, Muranen T, Tao J, Campos AB, Rodon J, Ibrahim YH, Serra V, Rodrik-Outmezguine V, Hazra S, Singh S, Kim P, Quadt C, Liu M, Huang A, Rosen N, Engelman JA, Scaltriti M, Baselga J. Elkabets M, et al. Sci Transl Med. 2013 Jul 31;5(196):196ra99. doi: 10.1126/scitranslmed.3005747. Sci Transl Med. 2013. PMID: 23903756 Free PMC article.
References
- Bader AG, Kang S, Zhao L, Vogt PK. Oncogenic PI3K deregulates transcription and translation. Nat Rev Cancer. 2005;5:921–929. - PubMed
- Bachman KE, et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004;3:772–775. - PubMed
- Broderick DK, et al. Mutations of PIK3CA in anaplastic oligodendrogliomas, high-grade astrocytomas, and medulloblastomas. Cancer Res. 2004;64:5048–5050. - PubMed
- Campbell IG, et al. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678–7681. - PubMed
- Hartmann C, et al. PIK3CA mutations in glioblastoma multiforme. Acta Neuropathol. 2005;109:639–642. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous