A role for autophagy in the extension of lifespan by dietary restriction in C. elegans - PubMed (original) (raw)
A role for autophagy in the extension of lifespan by dietary restriction in C. elegans
Malene Hansen et al. PLoS Genet. 2008 Feb.
Abstract
In many organisms, dietary restriction appears to extend lifespan, at least in part, by down-regulating the nutrient-sensor TOR (Target Of Rapamycin). TOR inhibition elicits autophagy, the large-scale recycling of cytoplasmic macromolecules and organelles. In this study, we asked whether autophagy might contribute to the lifespan extension induced by dietary restriction in C. elegans. We find that dietary restriction and TOR inhibition produce an autophagic phenotype and that inhibiting genes required for autophagy prevents dietary restriction and TOR inhibition from extending lifespan. The longevity response to dietary restriction in C. elegans requires the PHA-4 transcription factor. We find that the autophagic response to dietary restriction also requires PHA-4 activity, indicating that autophagy is a transcriptionally regulated response to food limitation. In spite of the rejuvenating effect that autophagy is predicted to have on cells, our findings suggest that autophagy is not sufficient to extend lifespan. Long-lived daf-2 insulin/IGF-1 receptor mutants require both autophagy and the transcription factor DAF-16/FOXO for their longevity, but we find that autophagy takes place in the absence of DAF-16. Perhaps autophagy is not sufficient for lifespan extension because although it provides raw material for new macromolecular synthesis, DAF-16/FOXO must program the cells to recycle this raw material into cell-protective longevity proteins.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
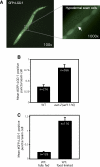
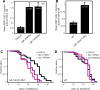
Figure 1. Dietary Restriction Increases the Level of Autophagy
LGG-1::GFP-positive puncta labeling autophagic membranes [22] were counted in wild-type or in food-limited animals. (A) Micrographs of eat-2(ad1116) L3 larvae expressing GFP-tagged lgg-1/LC3. Arrow indicates autophagic focus. Magnification is indicated. (B) Average number of LGG-1::GFP-containing puncta in eat-2(ad1116) mutants and N2 wild-type animals (WT), p < 0.0001. (C) Average number of LGG-1::GFP-containing puncta in N2 wild-type, food-restricted animals grown in liquid media (WT, food limited) and N2 wild-type animals grown in liquid with a higher concentration of bacteria (WT, fully fed), p < 0.0001; see Methods. Between three and ten seam cells were counted in each of 20–40 animals using high-power microscopy and averaged. n, total number of seam cells observed. Error bars: ±SEM. _p_-Values were calculated as unpaired, two-tailed t-test. Animals were raised at 20 °C. Please see Table S1 for quantification of all data.
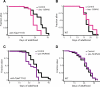
Figure 2. Inhibition of Genes Required for Autophagy Shortens the Long Lifespan of eat-2 Mutants
(A) Survival curves of eat-2(ad1116) animals fed either control bacteria or bacteria expressing bec-1 dsRNA during adulthood at 20 °C. Mean lifespan was 23.7 d for control and 19.6 d for bec-1 RNAi_, p_ < 0.0001, Log-rank (Mantel-Cox) test. This experiment was performed a total of six times, and bec-1 RNAi shortened the lifespan of eat-2 animals ∼15%–30%. Please see Table 1 for additional data. (B) Survival curves of N2 wild-type animals (WT) fed either control bacteria or bacteria expressing bec-1 dsRNA during adulthood at 20 °C. These assays were performed concurrently with the eat-2 mutant lifespan analysis in Figure 2A. Mean lifespan was 17.3 d for control and 18.9 d for bec-1 RNAi. p = 0.045, Log-rank (Mantel-Cox) test. Depletion of bec-1 did not significantly change the lifespan of N2 or sterile fer-15(b26); fem-1(hc17) animals in any of six experiments. Please see Table 1 for additional data. (C) Survival curves of eat-2(ad1116) animals fed either control bacteria or bacteria expressing vps-34 dsRNA during adulthood at 20 °C. Mean lifespan was 27.6 d for control and 22.8 d for vps-34 RNAi_, p_ = 0.0003, Log-rank (Mantel-Cox) test. This experiment was performed a total of four times. Please see Table 1 for additional data. (D) Survival curves of sterile fer-15(b26); fem-1(hc17) animals (WT) fed either control bacteria or bacteria expressing vps-34 dsRNA during adulthood at 20 °C. These assays were performed at the same time as the eat-2 lifespan analysis shown in Figure 2C. Mean lifespan was 21.5 d for control and 23.3 d for bec-1 RNAi. p = 0.14, Log-rank (Mantel-Cox) test. Depletion of vps-34 did not significantly change the lifespan of N2 or sterile fer-15(b26); fem-1(hc17) animals in each of six different experiments. Please see Table 1 for additional data.
Figure 3. Inhibition of the FOXA Transcription Factor pha-4 Decreases Autophagy in eat-2 and rab-10 Mutants
(A) Average number of LGG-1::GFP-containing puncta in eat-2(ad1116) progeny of animals fed either control bacteria or bacteria expressing bec-1, vps-34 or pha-4 dsRNA their entire lives. p ≤ 0.0001 for bec-1, vps-34 and pha-4 RNAi treatments compared to control, respectively, unpaired, two-tailed t-test. n, total number of seam cells observed. Error bars: ±SEM. See Figure 1 for details. (B) Average number of LGG-1::GFP-containing puncta in rab-10(ok1494) progeny of animals fed either control bacteria or bacteria expressing pha-4 dsRNA their entire lives; p < 0.0001, unpaired, two-tailed t-test. n, total number of seam cells observed. Error bars: ±SEM. Please see Figure 1 for details and Table S1 for quantification of data. Feeding mutants for several generations with pha-4 dsRNA, inhibited development and sharply decreased the number of eggs laid ([46], data not shown). However, the loss of many embryos was unlikely to bias our findings, since many progeny of daf-2 mutants treated with pha-4 RNAi also died but those that did not exhibited a robust autophagy phenotype (Figure 6).
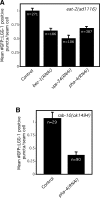
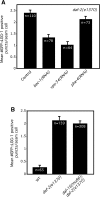
Figure 4. Animals with Low TOR-Pathway Activity Exhibit Increased Autophagy and Require the Autophagy-associated Gene bec-1 during Adulthood to Live Long
(A) Average number of LGG-1::GFP-containing puncta in let-363/TOR RNAi-arrested animals compared to N2 wild-type animals (WT) grown on control (vector-only) bacteria, p < 0.0001, unpaired, two-tailed t-test. n, total number of seam cells observed. Error bars: ±SEM. See Figure 1 for details. We were not able to detect increased LGG-1::GFP puncta in long-lived let-363(RNAi) adults; however, one generation of daf-2 RNAi, our positive control, did not significantly increase the number of foci in adults either (data not shown). Please see Table S1 for quantification of all data. (B) Average number of LGG-1::GFP-containing puncta in daf-15(m81)/unc-24(e138) heterozygotes (daf-15/+) compared to N2 wild-type animals (WT), p < 0.0001, unpaired, two-tailed t-test. n, total number of seam cells observed. Error bars: ±SEM. See Figure 1 for details. Please see Table S1 for quantification of all data. daf-15 encodes the TOR-binding partner Raptor. (C) Survival curves of daf-15(m81)/unc-24(e138) heterozygotes (daf-15/+, strain DR412) fed either control bacteria or bacteria expressing bec-1 dsRNA during adulthood at 20 °C. Mean lifespan: daf-15/+ animals grown on control RNAi-bacteria: 25.1 d, _daf-15/+_animals on bec-1 RNAi: 20.8 d, p = 0.0008, Log-rank (Mantel-Cox) test. The lifespan of daf-15/+ animals grown on bec-1 RNAi-bacteria during adulthood was measured again, yielding similar results; the lifespan of daf-15/+ animals was also measured three times following whole-life RNAi exposure. In these experiments, bec-1 RNAi generally shortened the mean lifespan of daf-15/+ animals to a greater extent than it shortened the lifespan of wild type. (We also attempted to perform double-RNAi experiments, in which animals were cultured on a 50:50 mixture of _let-363/_TOR and bec-1 [or control] RNAi bacteria. Although the trends we saw were in the expected direction, the effects produced by half-strength RNAi were small and not statistically significant [data not shown].) Please see Table 2 for additional data. (D) Survival curves of wild-type animals derived from strain DR412 (WT) fed either control bacteria or bacteria expressing bec-1 dsRNA throughout their whole life at 20 °C. These assays were performed concurrently with the daf-15/+ lifespan analysis shown in Figure 4C. WT grown on control RNAi-bacteria: 21.4 d, WT on bec-1 RNAi-bacteria: 21.1 d (p = 0.34), p between daf-15/+ and WT grown on control RNAi-bacteria, p < 0.0001, Log-rank (Mantel-Cox) test. Please see Table 2 for additional data.
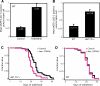
Figure 5. Inhibition of the Small GTPase rab-10 Increases Autophagy
(A) Average number of LGG-1::GFP-containing puncta in N2 wild-type animals (WT) fed either control bacteria or bacteria expressing daf-2 (as a control) or rab-10 dsRNA for two generations. p < 0.0001 for either daf-2 or rab-10 RNAi treatment compared to control RNAi treatment, unpaired, two-tailed t-test. n, total number of seam cells observed. Error bars: ±SEM. See Figure 1 for details. Please see Table S1 for quantification of all data. (B) Average number of LGG-1::GFP-containing puncta in rab-10(ok1494) mutants compared to N2 wild-type animals (WT), p < 0.0001, unpaired, two-tailed t-test. n, total number of seam cells observed. Error bars: ±SEM. Please see Figure 1 for details. Please see Table S1 for quantification of all data. (C) Survival curves of rab-10(ok1494) animals fed either control bacteria or bacteria expressing bec-1 or vps-34 dsRNA during adulthood at 20 °C. Mean lifespan was 27.9 d for control, 19.9 d for bec-1 RNAi, and 22.1 d for vps-34 RNAi_,_ all pair-wise comparisons to control, p < 0.0001, Log-rank (Mantel-Cox) test. This experiment was performed two times. Please see Table 3 for additional data. (D) Survival curves of N2 wild-type animals (WT) fed either control bacteria or bacteria expressing bec-1 or vps-34 dsRNA during adulthood at 20 °C. These assays were performed at the same time as the rab-10 lifespan analysis shown in Figure 5C. Mean lifespan was 22.9 d for control, 23.6 d for bec-1 RNAi, and 24.5 d for vps-34 RNAi, pair-wise comparison to control, p = 0.057 and p = 0.16, respectively, Log-rank (Mantel-Cox) test. Please see Table 1 for additional data.
Figure 6. Increased Autophagy in _daf-2/_insulin/IGF Receptor Mutants Is Independent of the daf-16/FOXO and pha-4/FOXA Transcription Factors
(A) Average number of LGG-1::GFP-containing puncta in daf-2(e1370) progeny of animals fed either control bacteria or bacteria expressing bec-1, vps-34, or pha-4 dsRNA for their entire lives. p < 0.0001 for bec-1 and vps-34 RNAi-bacteria compared to control RNAi-bacteria, respectively, p = 0.17 for pha-4 RNAi-bacteria compared to control RNAi-bacteria, unpaired, two-tailed t-test. n, total number of seam cells observed. Error bars: ±SEM. See Figure 1 for details. Feeding daf-2 mutants for several generations with pha-4 dsRNA sharply decreased the number of eggs laid (data not shown). The mean lifespan of daf-2(e1370) animals was shortened 12.5% by pha-4 RNAi ([28] and data not shown), and we measured an 11% decrease in puncta in daf-2(e1370) animals fed pha-4 RNAi. Even though this decrease was not statistically significant it remains possible that it relates to the small difference seen in lifespan. (B) Average number of LGG-1::GFP-containing puncta in daf-16(mu86); daf-2(e1370) double mutants compared to daf-2(e1370) animals; p = 0.50, unpaired, two-tailed t-test. N2 wild-type animals (WT) are shown for comparison. n, total number of seam cells observed. Error bars: ±SEM. See Figure 1 for details. The double mutant expressing the LGG-1 reporter had a mean lifespan similar to non-transgenic daf-16; daf-2 double mutants (data not shown, [65]). Please see Table S1 for quantification of all data.
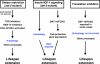
Figure 7. Model for the Role of Autophagy in Lifespan Extension by Dietary Restriction in C. elegans
In response to dietary restriction using the eat-2 mutation, TOR and RAB-10 activities fall, which triggers autophagy. Autophagy requires altered gene expression, since its appearance requires the transcription factor PHA-4. Autophagy is necessary but not sufficient for lifespan extension in long-lived daf-2 insulin/IGF-1-receptor mutants; instead, autophagy and the DAF-16/FOXO transcription factor are both required, independently, for lifespan extension. Inhibiting protein synthesis in well-fed animals may activate a distinct longevity pathway, since inhibiting protein synthesis in the context of dietary restriction produces a lifespan extension that requires autophagy gene function, whereas inhibiting protein synthesis in well-fed animals produces a lifespan extension that is independent of autophagy gene function. The sir-2 histone deacetylase is not in this diagram, as we [6] and several other groups [–68] have found that sir-2 deletion mutations do not prevent dietary restriction from increasing lifespan in C. elegans.
Similar articles
- Lifespan extension by suppression of autophagy genes in Caenorhabditis elegans.
Hashimoto Y, Ookuma S, Nishida E. Hashimoto Y, et al. Genes Cells. 2009 Jun;14(6):717-26. doi: 10.1111/j.1365-2443.2009.01306.x. Epub 2009 May 14. Genes Cells. 2009. PMID: 19469880 - Autophagy is required for dietary restriction-mediated life span extension in C. elegans.
Jia K, Levine B. Jia K, et al. Autophagy. 2007 Nov-Dec;3(6):597-9. doi: 10.4161/auto.4989. Epub 2007 Sep 6. Autophagy. 2007. PMID: 17912023 - The Target of Rapamycin pathway antagonizes pha-4/FoxA to control development and aging.
Sheaffer KL, Updike DL, Mango SE. Sheaffer KL, et al. Curr Biol. 2008 Sep 23;18(18):1355-64. doi: 10.1016/j.cub.2008.07.097. Curr Biol. 2008. PMID: 18804378 Free PMC article. - The search for DAF-16/FOXO transcriptional targets: approaches and discoveries.
Murphy CT. Murphy CT. Exp Gerontol. 2006 Oct;41(10):910-21. doi: 10.1016/j.exger.2006.06.040. Epub 2006 Aug 24. Exp Gerontol. 2006. PMID: 16934425 Review. - Longevity and heat stress regulation in Caenorhabditis elegans.
Muñoz MJ. Muñoz MJ. Mech Ageing Dev. 2003 Jan;124(1):43-8. doi: 10.1016/s0047-6374(02)00168-9. Mech Ageing Dev. 2003. PMID: 12618005 Review.
Cited by
- Deep Proteome Analysis Identifies Age-Related Processes in C. elegans.
Narayan V, Ly T, Pourkarimi E, Murillo AB, Gartner A, Lamond AI, Kenyon C. Narayan V, et al. Cell Syst. 2016 Aug;3(2):144-159. doi: 10.1016/j.cels.2016.06.011. Epub 2016 Jul 21. Cell Syst. 2016. PMID: 27453442 Free PMC article. - The Intersection of Aging, Longevity Pathways, and Learning and Memory in C. elegans.
Stein GM, Murphy CT. Stein GM, et al. Front Genet. 2012 Nov 26;3:259. doi: 10.3389/fgene.2012.00259. eCollection 2012. Front Genet. 2012. PMID: 23226155 Free PMC article. - Host-commensal interaction promotes health and lifespan in _Caenorhabditis elegans through the activation of HLH-30/TFEB-mediated autophagy.
Dinić M, Herholz M, Kačarević U, Radojević D, Novović K, Đokić J, Trifunović A, Golić N. Dinić M, et al. Aging (Albany NY). 2021 Mar 26;13(6):8040-8054. doi: 10.18632/aging.202885. Epub 2021 Mar 26. Aging (Albany NY). 2021. PMID: 33770762 Free PMC article. - ω-6 Polyunsaturated fatty acids extend life span through the activation of autophagy.
O'Rourke EJ, Kuballa P, Xavier R, Ruvkun G. O'Rourke EJ, et al. Genes Dev. 2013 Feb 15;27(4):429-40. doi: 10.1101/gad.205294.112. Epub 2013 Feb 7. Genes Dev. 2013. PMID: 23392608 Free PMC article. - Signaling pathways in mitochondrial dysfunction and aging.
Mammucari C, Rizzuto R. Mammucari C, et al. Mech Ageing Dev. 2010 Jul-Aug;131(7-8):536-43. doi: 10.1016/j.mad.2010.07.003. Epub 2010 Jul 24. Mech Ageing Dev. 2010. PMID: 20655326 Free PMC article. Review.
References
- Guarente L, Picard F. Calorie restriction–the SIR2 connection. Cell. 2005;120:473–482. - PubMed
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, et al. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. - PubMed
- Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. - PubMed
- Luong N, Davies CR, Wessells RJ, Graham SM, King MT, et al. Activated FOXO-mediated insulin resistance is blocked by reduction of TOR activity. Cell Metab. 2006;4:133–142. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T-32AG000278/AG/NIA NIH HHS/United States
- R01 AG024882/AG/NIA NIH HHS/United States
- T32 AG000278/AG/NIA NIH HHS/United States
- 5T90DK070135-02/DK/NIDDK NIH HHS/United States
- T90 DK070135/DK/NIDDK NIH HHS/United States
- 1 R01 AG024882/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous