Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin - PubMed (original) (raw)
. 2008 Mar 11;105(10):3945-50.
doi: 10.1073/pnas.0800135105. Epub 2008 Feb 28.
Michela Garofalo, Maria Paola Martelli, Roger Briesewitz, Lisheng Wang, Cecilia Fernandez-Cymering, Stefano Volinia, Chang-Gong Liu, Susanne Schnittger, Torsten Haferlach, Arcangelo Liso, Daniela Diverio, Marco Mancini, Giovanna Meloni, Robin Foa, Massimo F Martelli, Cristina Mecucci, Carlo M Croce, Brunangelo Falini
Affiliations
- PMID: 18308931
- PMCID: PMC2268779
- DOI: 10.1073/pnas.0800135105
Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin
Ramiro Garzon et al. Proc Natl Acad Sci U S A. 2008.
Erratum in
- Correction for Garzon et al., Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin.
[No authors listed] [No authors listed] Proc Natl Acad Sci U S A. 2024 Sep 3;121(36):e2415544121. doi: 10.1073/pnas.2415544121. Epub 2024 Aug 30. Proc Natl Acad Sci U S A. 2024. PMID: 39213182 Free PMC article. No abstract available. - Correction for Garzon et al., Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin.
[No authors listed] [No authors listed] Proc Natl Acad Sci U S A. 2024 Oct 22;121(43):e2419360121. doi: 10.1073/pnas.2419360121. Epub 2024 Oct 18. Proc Natl Acad Sci U S A. 2024. PMID: 39423245 Free PMC article. No abstract available.
Abstract
Acute myeloid leukemia (AML) carrying NPM1 mutations and cytoplasmic nucleophosmin (NPMc+ AML) accounts for about one-third of adult AML and shows distinct features, including a unique gene expression profile. MicroRNAs (miRNAs) are small noncoding RNAs of 19-25 nucleotides in length that have been linked to the development of cancer. Here, we investigated the role of miRNAs in the biology of NPMc+ AML. The miRNA expression was evaluated in 85 adult de novo AML patients characterized for subcellular localization/mutation status of NPM1 and FLT3 mutations using a custom microarray platform. Data were analyzed by using univariate t test within BRB tools. We identified a strong miRNA signature that distinguishes NPMc+ mutated (n = 55) from the cytoplasmic-negative (NPM1 unmutated) cases (n = 30) and includes the up-regulation of miR-10a, miR-10b, several let-7 and miR-29 family members. Many of the down-regulated miRNAs including miR-204 and miR-128a are predicted to target several HOX genes. Indeed, we confirmed that miR-204 targets HOXA10 and MEIS1, suggesting that the HOX up-regulation observed in NPMc+ AML may be due in part by loss of HOX regulators-miRNAs. FLT3-ITD+ samples were characterized by up-regulation of miR-155. Further experiments demonstrated that the up-regulation of miR-155 was independent from FLT3 signaling. Our results identify a unique miRNA signature associated with NPMc+ AML and provide evidence that support a role for miRNAs in the regulation of HOX genes in this leukemia subtype. Moreover, we found that miR-155 was strongly but independently associated with FLT3-ITD mutations.
Conflict of interest statement
B.F. and C.M. have applied for a patent on clinical use of NPM1 mutants. The authors declare no other conflict of interest.
Figures
Fig. 1.
Hierarchical cluster analysis of NK-AML patients. Overview of two-way (genes against samples) hierarchical cluster (Euclidean distance) of 85 AML samples using the genes that varies the most between samples. Mean centered signal intensities of gene-expression are depicted by a log-transformed (2 scale). Color areas indicate relative expression of each gene with respect to the gene median expression (red above, green below the median value and black, samples with signal intensity to background of 2 or less). Some miRNAs are represented by more than one probe. As shown, the clustering is mainly determined by the presence of cytoplasmic nucleophosmin (NPMc+). Among the _NPMc_+ positive samples, we identified also many subgroups. Patients with _FLT3_-ITD+ segregated among the different clusters.
Fig. 2.
MiR-204 targets HOXA10 and MEIS1 in AML. (A) Western blotting showing HOXA10 protein using whole cell lysates from MEG-01 and OCI-AML3 cell lines transfected with scrambled oligonucleotides or miR-139, miR-128a, and miR-204. The blots were stripped and reprobed with β-actin for loading control. (B) Western blotting showing MEIS1 protein expression in OCI-AML3 cell lines transfected with scrambled oligonucleotides or sense miR-204 and miR-101. (C) Western blotting showing HOXA10 and MEIS1 protein expression in MEG-01 and K562 cells transfected with scrambled oligonucleotides or antisense oligonucleotides against miR-204. The numbers below the immunoblot images represent the intensity of the HOXA-10/MEIS bands relative to the β-actin expression. (D) Relative luciferase activity in MEG01 cells transiently cotransfected with the luciferase reporter vector containing the 3′ UTR of MEIS1 with miR-204 or scrambled oligonucleotides. The results are presented as a fold difference in the luciferase/Renilla ratios with respect to the luciferase reporter constructs transfected with scrambled oligonucleotides.
Fig. 3.
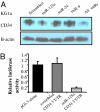
MiR-125a targets CD34. (A) Western blotting showing CD34 protein expression in KG1a cells 48 h after transfection with scrambled oligonucleotides or sense miR-125a, mIR-24, miR-9, or all combined oligonucleotides (B) Relative luciferase activity in KG1a cells transiently cotransfected with the luciferase reporter vector containing the 3′ UTR of CD34 predicted to interact with miR-125a or scrambled oligonucleotides. The results are presented as a fold difference in the luciferase/Renilla ratios with respect to the empty reporter (pGl3 alone).
Fig. 4.
MiR-155 is overexpressed in _FLT3_-ITD+ AML. (A) Average miR-155 expression in 32 NK-AML patients with _FLT3_-ITD+ (n = 10) and _FLT3_-wt (n = 22) after normalization with 18s and 2ΔCt calculations. The average miR-155 expression values between the two groups were compared by using t test. (B) IP/Western blot. MV4-11 cells were incubated for 24 and 48 h with SU14813 (300 nM) or DMSO (vehicle control). Lysates were generated and an immunoprecipitation was performed with an antibody for FLT3. SDS/PAGE was performed, followed by a Western blot with an antibody for phospho-tyrosine. Subsequently, the blot was stripped and reprobed with anti-FLT3. (C) Northern blotting of miR-155 in MV4-11 cells incubated for 24 and 48 h with SU14813 (300 nM) or DMSO (vehicle control). Total RNA was obtained with TRIzol and Northern blotting was performed with a probe antisense to miR-155. The blot was stripped and reprobed with U6 for loading control. (D) IP/Western blot. BaF3 and BaF3 _FLT3_-ITD cells were incubated for 24 h with SU14813 (300 nM) or DMSO (vehicle control). Lysates were generated and an immunoprecipitation was performed with an antibody for FLT3. SDS/PAGE was performed, followed by a Western blot with an antibody for phospho-tyrosine. Subsequently, the blot was stripped and reprobed with anti-FLT3. (E) Northern blotting for miR-155 in BaF3 and Baf3 _FLT3_-ITD clones incubated for 24 and 48 h with SU14813 or DMSO (control) as described. The blot was stripped and reprobed with U6 for loading control.
Similar articles
- Targeting Chromatin Regulators Inhibits Leukemogenic Gene Expression in NPM1 Mutant Leukemia.
Kühn MW, Song E, Feng Z, Sinha A, Chen CW, Deshpande AJ, Cusan M, Farnoud N, Mupo A, Grove C, Koche R, Bradner JE, de Stanchina E, Vassiliou GS, Hoshii T, Armstrong SA. Kühn MW, et al. Cancer Discov. 2016 Oct;6(10):1166-1181. doi: 10.1158/2159-8290.CD-16-0237. Epub 2016 Aug 17. Cancer Discov. 2016. PMID: 27535106 Free PMC article. - Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: a Cancer and Leukemia Group B Study.
Marcucci G, Maharry K, Radmacher MD, Mrózek K, Vukosavljevic T, Paschka P, Whitman SP, Langer C, Baldus CD, Liu CG, Ruppert AS, Powell BL, Carroll AJ, Caligiuri MA, Kolitz JE, Larson RA, Bloomfield CD. Marcucci G, et al. J Clin Oncol. 2008 Nov 1;26(31):5078-87. doi: 10.1200/JCO.2008.17.5554. Epub 2008 Sep 22. J Clin Oncol. 2008. PMID: 18809607 Free PMC article. - Molecular synergy underlies the co-occurrence patterns and phenotype of _NPM1_-mutant acute myeloid leukemia.
Dovey OM, Cooper JL, Mupo A, Grove CS, Lynn C, Conte N, Andrews RM, Pacharne S, Tzelepis K, Vijayabaskar MS, Green P, Rad R, Arends M, Wright P, Yusa K, Bradley A, Varela I, Vassiliou GS. Dovey OM, et al. Blood. 2017 Oct 26;130(17):1911-1922. doi: 10.1182/blood-2017-01-760595. Epub 2017 Aug 23. Blood. 2017. PMID: 28835438 Free PMC article. - Nucleophosmin (NPM1) mutations in adult and childhood acute myeloid leukaemia: towards definition of a new leukaemia entity.
Rau R, Brown P. Rau R, et al. Hematol Oncol. 2009 Dec;27(4):171-81. doi: 10.1002/hon.904. Hematol Oncol. 2009. PMID: 19569254 Free PMC article. Review. - Acute myeloid leukemia carrying cytoplasmic/mutated nucleophosmin (NPMc+ AML): biologic and clinical features.
Falini B, Nicoletti I, Martelli MF, Mecucci C. Falini B, et al. Blood. 2007 Feb 1;109(3):874-85. doi: 10.1182/blood-2006-07-012252. Epub 2006 Sep 28. Blood. 2007. PMID: 17008539 Review.
Cited by
- A polymorphism in the 3'-untranslated region of the NPM1 gene causes illegitimate regulation by microRNA-337-5p and correlates with adverse outcome in acute myeloid leukemia.
Cheng CK, Kwan TK, Cheung CY, Ng K, Liang P, Cheng SH, Chan NP, Ip RK, Wong RS, Lee V, Li CK, Yip SF, Ng MH. Cheng CK, et al. Haematologica. 2013 Jun;98(6):913-7. doi: 10.3324/haematol.2012.073015. Epub 2012 Oct 12. Haematologica. 2013. PMID: 23065518 Free PMC article. - The miRacle in Pancreatic Cancer by miRNAs: Tiny Angels or Devils in Disease Progression.
Hawa Z, Haque I, Ghosh A, Banerjee S, Harris L, Banerjee SK. Hawa Z, et al. Int J Mol Sci. 2016 May 26;17(6):809. doi: 10.3390/ijms17060809. Int J Mol Sci. 2016. PMID: 27240340 Free PMC article. Review. - miRNA: A Promising Therapeutic Target in Cancer.
Menon A, Abd-Aziz N, Khalid K, Poh CL, Naidu R. Menon A, et al. Int J Mol Sci. 2022 Sep 29;23(19):11502. doi: 10.3390/ijms231911502. Int J Mol Sci. 2022. PMID: 36232799 Free PMC article. Review. - Targeting microRNAs in cancer: rationale, strategies and challenges.
Garzon R, Marcucci G, Croce CM. Garzon R, et al. Nat Rev Drug Discov. 2010 Oct;9(10):775-89. doi: 10.1038/nrd3179. Nat Rev Drug Discov. 2010. PMID: 20885409 Free PMC article. Review. - microRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia.
Han YC, Park CY, Bhagat G, Zhang J, Wang Y, Fan JB, Liu M, Zou Y, Weissman IL, Gu H. Han YC, et al. J Exp Med. 2010 Mar 15;207(3):475-89. doi: 10.1084/jem.20090831. Epub 2010 Mar 8. J Exp Med. 2010. PMID: 20212066 Free PMC article.
References
- Lowenberg B, Downing JR, Burnett A. Acute Myeloid leukemia. N Engl J Med. 1999;341:1051–1062. - PubMed
- Burnett AK. Current controversies: Which patients with acute myeloid leukemia should receive bone marrow transplantation? An adult theater's view. Br J Haematol. 2002;118:357–364. - PubMed
- Kiyoi H, et al. Prognostic implication of FLT3 and N-RAS gene mutations in acute myeloid leukemia. Blood. 1999;93:3074–3080. - PubMed
- Shih LY, et al. Internal tandem duplication of FLT3 in relapsed acute myeloid leukemia: A comparative analysis of bone marrow samples from 108 adult patients at diagnosis and relapse. Blood. 2000;100:2387–2392. - PubMed
- Frohling S, et al. CEBPA mutations in younger adults with acute myeloid leukemia and normal cytogenetics: Prognostic relevance and analysis of cooperating mutations. J Clin Oncol. 2004;22:624–633. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous