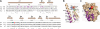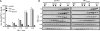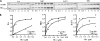Binding of yeast frataxin to the scaffold for Fe-S cluster biogenesis, Isu - PubMed (original) (raw)
Binding of yeast frataxin to the scaffold for Fe-S cluster biogenesis, Isu
Tao Wang et al. J Biol Chem. 2008.
Abstract
Friedreich ataxia is caused by reduced activity of frataxin, a conserved iron-binding protein of the mitochondrial matrix, thought to supply iron for formation of Fe-S clusters on the scaffold protein Isu. Frataxin binds Isu in an iron-dependent manner in vitro. However, the biological relevance of this interaction and whether in vivo the interaction between frataxin and Isu is mediated by adaptor proteins is a matter of debate. Here, we report that alterations of conserved, surface-exposed residues of yeast frataxin, which have deleterious effects on cell growth, impair Fe-S cluster biogenesis and interaction with Isu while altering neither iron binding nor oligomerization. Our results support the idea that the surface of the beta-sheet, adjacent to the acidic, iron binding ridge, is important for interaction of Yfh1 with the Fe-S cluster scaffold and point to a critical role for frataxin in Fe-S cluster biogenesis.
Figures
FIGURE 1.
Yfh1 structure. A, ClustalX alignment of sequences corresponding to the α1-α2 region of Yfh1. S. cerevisiae, Yfh1 (Sc); Homo sapiens, frataxin (Hs);Escherichia coli, CyaY (Ec). Secondary structural elements and numbering referring to Yfh1 are above the sequences. Iron binding residues from Ref. (purple), Asn-122 (green), Lys-123 (blue), Gln-124 (red) are indicated. Identical (*) and highly similar (:) residues are indicated. Ribbon (B) and space-filled Yfh1 (Protein Data Bank accession code 2ga5) (C), generated using PyMOL (29). Color coding as in_A_. The N-terminal residues (52-67) are not shown.
FIGURE 2.
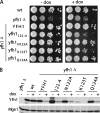
Growth of YFH1 mutants. A, 10-fold serial dilutions of cell suspensions of WT, yfh1_Δ, and_yfh1_Δ transformed with plasmids carrying the indicated tetO-regulatable WT or mutant YFH1 were plated on rich glucose medium containing (+) or lacking (-) doxycycline (dox).yfhN122A/K123T/Q124A, (yfh1122-4). Plates were incubated at 30 °C for 2 days. B, cells grown as in_A were harvested and subjected to SDS-PAGE followed by immunoblot analysis using Yfh1 and, as a loading control, Mge1-specific antibodies.
FIGURE 3.
Iron binding and oligomerization. A, purified Yfh1 proteins were incubated with ferrous iron at the indicated protein to iron ratios and then split into two equal parts and subjected to iron quantitation spectroscopically (see “Experimental Procedures” for details).Bars represent the means ± S.E. of at least three independent reactions. B, gel filtration chromatography. Fractions were resolved by SDS-PAGE and stained with SYPRO-Ruby. Positions of thymoglobulin (669 kDa), aldolase (158 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa), and ribonuclease A (13.7 kDa) are indicated.
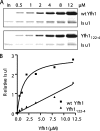
FIGURE 4.
Interaction with Isu and other Fe-S cluster assembly proteins in mitochondria extracts. A, indicated concentrations of His-tagged Yfh1 or Yfh122-4 were incubated with a mitochondrial extract. Proteins pulled down by Ni-NTA resins were detected by immunoblot using antibodies against indicated proteins after resolution by SDS-PAGE. Left lane, 5% of total input (In). Similar experiments were done a minimum of three times; a representative experiment is shown. The band indicated by the asterisk (*) is a cross-reacting protein, unrelated to Isu. B, immunoblot signals were quantitated by densitometry and plotted against Yfh1 protein concentrations. 5% input was set as 1. All curves were plotted in Prism using a single binding hyperbola to fit data, except that Isu/Yfh122-4 was fitted by linear regression.
FIGURE 5.
Yfh1 interaction with Isu and Isd11/Nfs1 in mitochondria extracts with up-regulated expression level of Isu, Ids11, or Nfs1. A, expression level of Isu, Nfs1, and Isd11 in mitochondrial samples. N, normal expression; overexpression of Isu1 (U), Nfs1 (S), Isd11 (D). Mge1 serves as a loading control. B, purified His-tagged WT Yfh1 or Yfh122-4 (final concentration of ∼0.5 μ
m
) was added to the indicated mitochondrial extracts. Protein complexes pulled down by Ni-NTA resin were analyzed by immunoblot analysis using antibodies against the indicated proteins. Mge1 serves as a negative control. 5% input (In); WT Yfh1 (WT), Yfh1122-4 (122-4), control, no Yfh1 proteins were added to the reactions (C).
FIGURE 6.
Interaction of purified Yfh1 and Isu1. A, 5 μ
m
Isu1 and His-tagged Yfh1 or Yfh122-4 at the concentrations indicated were preincubated to allow complex formation. Ni-NTA resins were added to pull down the complexes that were analyzed by SDS-PAGE and SYPRO-Ruby staining. Leftmost lane, 20% Isu1 input. B, bound Isu1 was quantitated by densitometry. 20% input Isu1 was set as 1. Values were plotted in Prism using a single binding hyperbola to fit data obtained for wild type Yfh1 and linear regression to fit Yfh122-4 data.
FIGURE 7.
Characterization of FRDA-associated _yfh1_N122K. A and B, 10-fold serial dilutions of _yfh1_Δ (A) or _yfh1_Δ/_isu1_Δ (B) transformed with plasmids carrying tetO-regulatable YFH1, yfh1N122K, or yfh1122-4 were spotted on rich glucose medium containing (+) or lacking (-) doxycycline and incubated at 30 °C for 2 days. A, right panel, immunoblot analysis of lysates from plates using Yfh1-specific antibody. C, Yfh1N122K interaction with Isu and other Fe-S cluster assembly proteins in mitochondria extracts. Indicated His-tagged Yfh1 were incubated with mitochondrial extract; proteins pulled down by Ni-NTA resins were detected using antibodies against the indicated proteins after SDS-PAGE. 5% of input (In); control, no Yfh1 protein added to reactions (C).
Similar articles
- Mutation in the Fe-S scaffold protein Isu bypasses frataxin deletion.
Yoon H, Golla R, Lesuisse E, Pain J, Donald JE, Lyver ER, Pain D, Dancis A. Yoon H, et al. Biochem J. 2012 Jan 1;441(1):473-80. doi: 10.1042/BJ20111637. Biochem J. 2012. PMID: 21936771 Free PMC article. - Binding of the chaperone Jac1 protein and cysteine desulfurase Nfs1 to the iron-sulfur cluster scaffold Isu protein is mutually exclusive.
Majewska J, Ciesielski SJ, Schilke B, Kominek J, Blenska A, Delewski W, Song JY, Marszalek J, Craig EA, Dutkiewicz R. Majewska J, et al. J Biol Chem. 2013 Oct 4;288(40):29134-42. doi: 10.1074/jbc.M113.503524. Epub 2013 Aug 14. J Biol Chem. 2013. PMID: 23946486 Free PMC article. - Frataxin directly stimulates mitochondrial cysteine desulfurase by exposing substrate-binding sites, and a mutant Fe-S cluster scaffold protein with frataxin-bypassing ability acts similarly.
Pandey A, Gordon DM, Pain J, Stemmler TL, Dancis A, Pain D. Pandey A, et al. J Biol Chem. 2013 Dec 27;288(52):36773-86. doi: 10.1074/jbc.M113.525857. Epub 2013 Nov 11. J Biol Chem. 2013. PMID: 24217246 Free PMC article. - Frataxin and mitochondrial FeS cluster biogenesis.
Stemmler TL, Lesuisse E, Pain D, Dancis A. Stemmler TL, et al. J Biol Chem. 2010 Aug 27;285(35):26737-26743. doi: 10.1074/jbc.R110.118679. Epub 2010 Jun 3. J Biol Chem. 2010. PMID: 20522547 Free PMC article. Review. - Mechanisms of Mitochondrial Iron-Sulfur Protein Biogenesis.
Lill R, Freibert SA. Lill R, et al. Annu Rev Biochem. 2020 Jun 20;89:471-499. doi: 10.1146/annurev-biochem-013118-111540. Epub 2020 Jan 14. Annu Rev Biochem. 2020. PMID: 31935115 Review.
Cited by
- Crystal Structure of Bacillus subtilis Cysteine Desulfurase SufS and Its Dynamic Interaction with Frataxin and Scaffold Protein SufU.
Blauenburg B, Mielcarek A, Altegoer F, Fage CD, Linne U, Bange G, Marahiel MA. Blauenburg B, et al. PLoS One. 2016 Jul 6;11(7):e0158749. doi: 10.1371/journal.pone.0158749. eCollection 2016. PLoS One. 2016. PMID: 27382962 Free PMC article. - Pharmacological screening using an FXN-EGFP cellular genomic reporter assay for the therapy of Friedreich ataxia.
Li L, Voullaire L, Sandi C, Pook MA, Ioannou PA, Delatycki MB, Sarsero JP. Li L, et al. PLoS One. 2013;8(2):e55940. doi: 10.1371/journal.pone.0055940. Epub 2013 Feb 13. PLoS One. 2013. PMID: 23418481 Free PMC article. - Molecular details of the yeast frataxin-Isu1 interaction during mitochondrial Fe-S cluster assembly.
Cook JD, Kondapalli KC, Rawat S, Childs WC, Murugesan Y, Dancis A, Stemmler TL. Cook JD, et al. Biochemistry. 2010 Oct 12;49(40):8756-65. doi: 10.1021/bi1008613. Epub 2010 Sep 14. Biochemistry. 2010. PMID: 20815377 Free PMC article. - Metal acquisition and availability in the mitochondria.
Atkinson A, Winge DR. Atkinson A, et al. Chem Rev. 2009 Oct;109(10):4708-21. doi: 10.1021/cr900006y. Chem Rev. 2009. PMID: 19522505 Free PMC article. Review. No abstract available.
References
- Campuzano, V., Montermini, L., Molto, M. D., Pianese, L., Cossee, M., Cavalcanti, F., Monros, E., Rodius, F., Duclos, F., Monticelli, A., Zara, F., Canizares, J., Koutnikova, H., Bidichandani, S. I., Gellera, C., Brice, A., Trouillas, P., De Michele, G., Filla, A., De Frutos, R., Palau, F., Patel, P. I., Di Donato, S., Mandel, J. L., Cocozza, S., Koenig, M., and Pandolfo, M. (1996) Science 271 1423-1427 - PubMed
- Cossee, M., Durr, A., Schmitt, M., Dahl, N., Trouillas, P., Allinson, P., Kostrzewa, M., Nivelon-Chevallier, A., Gustavson, K. H., Kohlschutter, A., Muller, U., Mandel, J. L., Brice, A., Koenig, M., Cavalcanti, F., Tammaro, A., De Michele, G., Filla, A., Cocozza, S., Labuda, M., Montermini, L., Poirier, J., and Pandolfo, M. (1999) Ann. Neurol. 45 200-206 - PubMed
- Puccio, H., and Koenig, M. (2000) Hum. Mol. Genet. 9 887-892 - PubMed
- Babcock, M., de Silva, D., Oaks, R., Davis-Kaplan, S., Jiralerspong, S., Montermini, L., Pandolfo, M., and Kaplan, J. (1997) Science 276 1709-1712 - PubMed
- Rotig, A., de Lonlay, P., Chretien, D., Foury, F., Koenig, M., Sidi, D., Munnich, A., and Rustin, P. (1997) Nat. Genet. 17 215-217 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous