A polycomb group protein, PHF1, is involved in the response to DNA double-strand breaks in human cell - PubMed (original) (raw)
A polycomb group protein, PHF1, is involved in the response to DNA double-strand breaks in human cell
Zehui Hong et al. Nucleic Acids Res. 2008 May.
Abstract
DNA double-strand breaks (DSBs) represent the most toxic DNA damage arisen from endogenous and exogenous genotoxic stresses and are known to be repaired by either homologous recombination or nonhomologous end-joining processes. Although many proteins have been identified to participate in either of the processes, the whole processes still remain elusive. Polycomb group (PcG) proteins are epigenetic chromatin modifiers involved in gene silencing, cancer development and the maintenance of embryonic and adult stem cells. By screening proteins responding to DNA damage using laser micro-irradiation, we found that PHF1, a human homolog of Drosophila polycomb-like, Pcl, protein, was recruited to DSBs immediately after irradiation and dissociated within 10 min. The accumulation at DSBs is Ku70/Ku80-dependent, and knockdown of PHF1 leads to X-ray sensitivity and increases the frequency of homologous recombination in HeLa cell. We found that PHF1 interacts physically with Ku70/Ku80, suggesting that PHF1 promotes nonhomologous end-joining processes. Furthermore, we found that PHF1 interacts with a number of proteins involved in DNA damage responses, RAD50, SMC1, DHX9 and p53, further suggesting that PHF1, besides the function in PcG, is involved in genome maintenance processes.
Figures
Figure 1.
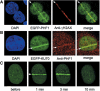
Recruitment of PHF1 to the sites irradiated with 405 nm laser. HeLa cells were sensitized with BrdU and micro-irradiated. Five minutes later, the cells were photographed (for GFP-tagged protein) or fixed and stained with antibody. EGFP-tagged PHF1 is recruited to laser-irradiated sites and co-localizes with γH2AX. (A) Accumulation of GFP-tagged PHF1 (EGFP-PHF1) and γH2AX. (B) Accumulation of EGFP-KU70 co-localized with endogenous PHF1. (C) Accumulation and dissociation of GFP-PHF1 at laser-irradiated sites in HeLa cells. Arrows indicate the sites of irradiation.
Figure 2.
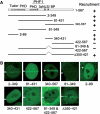
PHF1 is recruited to laser-irradiated sites via two independent domains of the N-terminus Tudor domain and the central region (aa 340–431). (A) Schematic presentation of PHF1 domains (top) and deletion mutants (left) and the results of recruitment experiments (right). (B) Live cell imaging of irradiated HeLa cells expressing GFP-tagged deletion mutants of PHF1. Arrows indicate the sites of irradiation.
Figure 3.
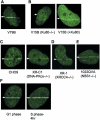
PHF1 is recruited to DSBs in dependence on Ku70/Ku80. (A) EGFP-PHF1 accumulates at laser-irradiated site in wild-type V79B. (B) EGFP-PHF1 does not accumulate at irradiation sites in V15B cell (Ku80-deficient) but do in V15B cell (+His-Ku80). (C) EGFP-PHF1 accumulates at irradiation sites both in DNA-Pkcs-deficient and proficient CHO cells. (D) EGFP-PHF1 accumulates at irradiation sites in XR-1 cell (XRCC4-deficient). (E) EGFP-PHF1 accumulates at irradiation sites in 1022QVA (NBS1-deficient). (F) HeLa cells were synchronized to G1- and G2/S-phase by thymidine and hydroxyurea treatment. PHF1 accumulates at irradiation sites in HeLa cells both in G1 phase and S phase. Arrows indicate the sites of irradiation.
Figure 4.
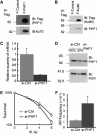
PHF1 is associated physically with Ku70/Ku80 and may be involved in NHEJ. (A) pcDNA5/FRT vector expressing FLAG-HA tagged PHF1 was transfected into Flp-In-293 cells, and a clone stably expressing FLAG-HA-PHF1 (F-PHF1) was isolated and used for coimmunoprecipitation by anti-FLAG M2 affinity gel. A clone stably expressing FLAG-HA tag alone (F-Control) was used as negative control. (B) pcDNA5/FRT vector expressing FLAG-HA tagged Ku80 was transfected into Flp-In-293 cells, and a clone stably expressing FLAG-HA-PHF1 (F-Ku80) was isolated and used for coimmunoprecipitation by anti-FLAG M2 affinity gel. (C) Real-time PCR analysis of a PHF1 stable knockdown cell line and a parallel mock knockdown cell line. HPRT was used as the control for normalization in the real-time PCR. (D) Immunoblotting analysis of PHF1 stable knockdown cell line and parallel mock knockdown cell line. Actin serves as a loading control for immunoblotting. (E) PHF1 knockdown cell showed increased sensitivity to X-Ray. Cells were plated in 6 cm dishes and irradiated with the indicated doses of ionizing irradiation. (F) Recombination frequency in HeLa cells after knockdown of PHF1. Error bars represent standard errors.
Figure 5.
Analysis of PHF1-associated proteins in HeLa cell by proteomics. (A) Putative PHF1-binding proteins identified by coimmunoprecipitation and mass spectrometry. Among identified proteins by mass spectrometry, those related to DNA damage response and PcG are indicated. Flp-In-293 cells stably expressing FLAG-HA tagged PHF1 was used for immunoprecipitation for FLAG. Flp-In-293 cells expressing FLAG-HA-tag alone was used as negative control. (B) Candidate interacting proteins identified by nanoLC/MS/MS were further confirmed using antibodies against SMC1, p53 and p46 by immunoblotting. Antibodies (IB) are indicated on the right.
Similar articles
- Polycomb group protein PHF1 regulates p53-dependent cell growth arrest and apoptosis.
Yang Y, Wang C, Zhang P, Gao K, Wang D, Yu H, Zhang T, Jiang S, Hexige S, Hong Z, Yasui A, Liu JO, Huang H, Yu L. Yang Y, et al. J Biol Chem. 2013 Jan 4;288(1):529-39. doi: 10.1074/jbc.M111.338996. Epub 2012 Nov 13. J Biol Chem. 2013. PMID: 23150668 Free PMC article. - Accumulation of Ku80 proteins at DNA double-strand breaks in living cells.
Koike M, Koike A. Koike M, et al. Exp Cell Res. 2008 Mar 10;314(5):1061-70. doi: 10.1016/j.yexcr.2007.11.014. Epub 2007 Nov 28. Exp Cell Res. 2008. PMID: 18164703 - The ACF1 complex is required for DNA double-strand break repair in human cells.
Lan L, Ui A, Nakajima S, Hatakeyama K, Hoshi M, Watanabe R, Janicki SM, Ogiwara H, Kohno T, Kanno S, Yasui A. Lan L, et al. Mol Cell. 2010 Dec 22;40(6):976-87. doi: 10.1016/j.molcel.2010.12.003. Mol Cell. 2010. PMID: 21172662 - The Ku heterodimer: function in DNA repair and beyond.
Fell VL, Schild-Poulter C. Fell VL, et al. Mutat Res Rev Mutat Res. 2015 Jan-Mar;763:15-29. doi: 10.1016/j.mrrev.2014.06.002. Epub 2014 Jul 4. Mutat Res Rev Mutat Res. 2015. PMID: 25795113 Review. - The emerging role of Polycomb repressors in the response to DNA damage.
Vissers JH, van Lohuizen M, Citterio E. Vissers JH, et al. J Cell Sci. 2012 Sep 1;125(Pt 17):3939-48. doi: 10.1242/jcs.107375. J Cell Sci. 2012. PMID: 23104738 Review.
Cited by
- An aromatic cage is required but not sufficient for binding of Tudor domains of the Polycomblike protein family to H3K36me3.
Gatchalian J, Kingsley MC, Moslet SD, Rosas Ospina RD, Kutateladze TG. Gatchalian J, et al. Epigenetics. 2015;10(6):467-73. doi: 10.1080/15592294.2015.1042646. Epub 2015 Apr 29. Epigenetics. 2015. PMID: 25923537 Free PMC article. - Novel fusion of MYST/Esa1-associated factor 6 and PHF1 in endometrial stromal sarcoma.
Panagopoulos I, Micci F, Thorsen J, Gorunova L, Eibak AM, Bjerkehagen B, Davidson B, Heim S. Panagopoulos I, et al. PLoS One. 2012;7(6):e39354. doi: 10.1371/journal.pone.0039354. Epub 2012 Jun 22. PLoS One. 2012. PMID: 22761769 Free PMC article. - BMI1 confers radioresistance to normal and cancerous neural stem cells through recruitment of the DNA damage response machinery.
Facchino S, Abdouh M, Chatoo W, Bernier G. Facchino S, et al. J Neurosci. 2010 Jul 28;30(30):10096-111. doi: 10.1523/JNEUROSCI.1634-10.2010. J Neurosci. 2010. PMID: 20668194 Free PMC article. - Resveratrol reduced the detrimental effects of malondialdehyde on human endothelial cells.
Hassanpour M, Biray Avci Ç, Rahbarghazi R, Rezabakhsh A, Nourazarian A, Nabat E, Fathi F, Khaksar M. Hassanpour M, et al. J Cardiovasc Thorac Res. 2021;13(2):131-140. doi: 10.34172/jcvtr.2021.27. Epub 2021 Apr 24. J Cardiovasc Thorac Res. 2021. PMID: 34326967 Free PMC article. - Engrailed Homeoprotein Protects Mesencephalic Dopaminergic Neurons from Oxidative Stress.
Rekaik H, Blaudin de Thé FX, Fuchs J, Massiani-Beaudoin O, Prochiantz A, Joshi RL. Rekaik H, et al. Cell Rep. 2015 Oct 13;13(2):242-50. doi: 10.1016/j.celrep.2015.08.076. Epub 2015 Sep 24. Cell Rep. 2015. PMID: 26411690 Free PMC article.
References
- Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair. 2006;5:1021–1029. - PubMed
- Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 2001;27:247–254. - PubMed
- Mills KD, Ferguson DO, Alt FW. The role of DNA breaks in genomic instability and tumorigenesis. Immunol. Rev. 2003;194:77–95. - PubMed
- Valerie K, Povirk LF. Regulation and mechanisms of mammalian double-strand break repair. Oncogene. 2003;22:5792–5812. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous