Structural and enzymatic analysis of MshA from Corynebacterium glutamicum: substrate-assisted catalysis - PubMed (original) (raw)
Structural and enzymatic analysis of MshA from Corynebacterium glutamicum: substrate-assisted catalysis
Matthew W Vetting et al. J Biol Chem. 2008.
Abstract
The glycosyltransferase termed MshA catalyzes the transfer of N-acetylglucosamine from UDP-N-acetylglucosamine to 1-L-myo-inositol-1-phosphate in the first committed step of mycothiol biosynthesis. The structure of MshA from Corynebacterium glutamicum was determined both in the absence of substrates and in a complex with UDP and 1-L-myo-inositol-1-phosphate. MshA belongs to the GT-B structural family whose members have a two-domain structure with both domains exhibiting a Rossman-type fold. Binding of the donor sugar to the C-terminal domain produces a 97 degrees rotational reorientation of the N-terminal domain relative to the C-terminal domain, clamping down on UDP and generating the binding site for 1-L-myo-inositol-1-phosphate. The structure highlights the residues important in binding of UDP-N-acetylglucosamine and 1-L-myo-inositol-1-phosphate. Molecular models of the ternary complex suggest a mechanism in which the beta-phosphate of the substrate, UDP-N-acetylglucosamine, promotes the nucleophilic attack of the 3-hydroxyl group of 1-L-myo-inositol-1-phosphate while at the same time promoting the cleavage of the sugar nucleotide bond.
Figures
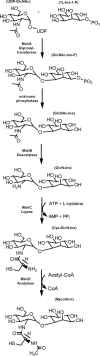
FIGURE 1.
Steps in the biosynthesis of mycothiol.
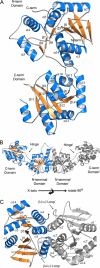
FIGURE 2.
Structure of APO CgMshA. A, ribbon diagram of an APO-CgMshA monomer (open conformation). Helices are in blue, strands are in_orange_, and coils are in gray. B, molecular dimer of CgMshA.C, illustration of the secondary structure involved in formation of the molecular dimer. In B and C, the 2-fold-related monomer is colored gray with secondary structure labeled with prime (′) designations (i.e. α1′ and β12′).
FIGURE 3.
Structure of the CgMshA UDP·1-l-Ins-1-P complex. A, final 2_Fo_ - Fc density for UDP and 1-
l
-Ins-1-P contoured at 1.0σ. B, ribbon diagram of a CgMshA monomer in complex with UDP and 1-
l
-Ins-1-P (closed conformation). UDP and 1-
l
-Ins-1-P are shown as sticks colored by atom type. C, molecular dimer of CgMshA after domain reorientation. Electron density for 1-
l
-Ins-1-P was sufficient only to fit in one subunit but was modeled in the dimer shown here for illustrative purposes. D, stereo illustration of DYNDOM rotation axis, relating the N- and C-terminal domain before and after binding of nucleoside. The APO structure is illustrated by the green trace, whereas the N- and C-terminal domains of the ternary complex are colored_blue_ and cyan, respectively. α12 is colored_maroon_ in both structures to help in orientation, whereas the hinge residues (196–197 and 386–392) are colored red.
FIGURE 4.
Interactions in the ternary complex. Illustrations of the interaction of CgMshA with the UDP-GlcNAc model and 1-
l
-Ins-1-P are shown. A, Chemdraw diagram. Interactions shown with UDP (maroon) and 1-
l
-Ins-1-P (blue) are those observed in the ternary complex, whereas those with GlcNAc (red) are those found in the molecular model. B, stereo stick diagram.1-
l
-Ins-1-P carbons are colored yellow, and UDP-GlcNAc carbons are colored green. Important hydrogen bonding interactions are shown as gray dotted lines, and the interaction of the 3-OH of 1-
l
-Ins-1-P with the UDP-GlcNAc at the site of reaction chemistry is shown as a cyan dotted line. C, proposed SNi-substrate-assisted catalysis mechanism, featuring oxocarbenium-ion like transition state, with asymmetric phosphor bond breakage, and glycosidic bond formation.
Similar articles
- In Vitro and In Silico Explorations of the Protein Conformational Changes of Corynebacterium glutamicum MshA, a Model Retaining GT-B Glycosyltransferase.
Hassan BA, Milicaj J, Tyson M, Karki R, Sham YY, Frantom PA, Taylor EA. Hassan BA, et al. Biochemistry. 2024 Apr 2;63(7):939-951. doi: 10.1021/acs.biochem.3c00561. Epub 2024 Mar 20. Biochemistry. 2024. PMID: 38507812 - Biochemistry of the initial steps of mycothiol biosynthesis.
Newton GL, Ta P, Bzymek KP, Fahey RC. Newton GL, et al. J Biol Chem. 2006 Nov 10;281(45):33910-20. doi: 10.1074/jbc.M604724200. Epub 2006 Aug 28. J Biol Chem. 2006. PMID: 16940050 - HDX-MS Reveals Substrate-Dependent, Localized EX1 Conformational Dynamics in the Retaining GT-B Glycosyltransferase, MshA.
Karki R, Hennek JT, Chen W, Frantom PA. Karki R, et al. Biochemistry. 2023 Sep 5;62(17):2645-2657. doi: 10.1021/acs.biochem.3c00338. Epub 2023 Aug 17. Biochemistry. 2023. PMID: 37589157 - UDP-(5F)-GlcNAc acts as a slow-binding inhibitor of MshA, a retaining glycosyltransferase.
Frantom PA, Coward JK, Blanchard JS. Frantom PA, et al. J Am Chem Soc. 2010 May 19;132(19):6626-7. doi: 10.1021/ja101231a. J Am Chem Soc. 2010. PMID: 20411981 Free PMC article. - Nucleotide substrate recognition by UDP-N-acetylglucosamine acyltransferase (LpxA) in the first step of lipid A biosynthesis.
Ulaganathan V, Buetow L, Hunter WN. Ulaganathan V, et al. J Mol Biol. 2007 Jun 1;369(2):305-12. doi: 10.1016/j.jmb.2007.03.039. Epub 2007 Mar 21. J Mol Biol. 2007. PMID: 17434525
Cited by
- Conserved Conformational Hierarchy across Functionally Divergent Glycosyltransferases of the GT-B Structural Superfamily as Determined from Microsecond Molecular Dynamics.
Ramirez-Mondragon CA, Nguyen ME, Milicaj J, Hassan BA, Tucci FJ, Muthyala R, Gao J, Taylor EA, Sham YY. Ramirez-Mondragon CA, et al. Int J Mol Sci. 2021 Apr 28;22(9):4619. doi: 10.3390/ijms22094619. Int J Mol Sci. 2021. PMID: 33924837 Free PMC article. - Characterization of the N-acetyl-α-D-glucosaminyl l-malate synthase and deacetylase functions for bacillithiol biosynthesis in Bacillus anthracis.
Parsonage D, Newton GL, Holder RC, Wallace BD, Paige C, Hamilton CJ, Dos Santos PC, Redinbo MR, Reid SD, Claiborne A. Parsonage D, et al. Biochemistry. 2010 Sep 28;49(38):8398-414. doi: 10.1021/bi100698n. Biochemistry. 2010. PMID: 20799687 Free PMC article. - Towards Computationally Guided Design and Engineering of a Neisseria meningitidis Serogroup W Capsule Polymerase with Altered Substrate Specificity.
Paudel S, Wachira J, McCarthy PC. Paudel S, et al. Processes (Basel). 2021 Dec;9(12):2192. doi: 10.3390/pr9122192. Epub 2021 Dec 6. Processes (Basel). 2021. PMID: 37483532 Free PMC article. - Structural basis of the molecular ruler mechanism of a bacterial glycosyltransferase.
Ramírez AS, Boilevin J, Mehdipour AR, Hummer G, Darbre T, Reymond JL, Locher KP. Ramírez AS, et al. Nat Commun. 2018 Jan 31;9(1):445. doi: 10.1038/s41467-018-02880-2. Nat Commun. 2018. PMID: 29386647 Free PMC article. - Mycothiol biosynthesis is essential for ethionamide susceptibility in Mycobacterium tuberculosis.
Vilchèze C, Av-Gay Y, Attarian R, Liu Z, Hazbón MH, Colangeli R, Chen B, Liu W, Alland D, Sacchettini JC, Jacobs WR Jr. Vilchèze C, et al. Mol Microbiol. 2008 Sep;69(5):1316-29. doi: 10.1111/j.1365-2958.2008.06365.x. Epub 2008 Jul 21. Mol Microbiol. 2008. PMID: 18651841 Free PMC article.
References
- Demain, A. L. (1999) Appl. Microbiol. Biotechnol. 52 455-463 - PubMed
- Hermann, T. (2003) J. Biotechnol. 104 155-172 - PubMed
- Dye, C., Scheele, S., Dolin, P., Pathania, V., and Raviglione, M. C. (1999) J. Am. Med. Assoc. 282 677-686 - PubMed
- Britton, W. J., and Lockwood, D. N. (2004) Lancet 363 1209-1219 - PubMed
- Hand, C. E., and Honek, J. F. (2005) J. Nat. Prod. 68 293-308 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous