Oligosaccharyltransferase-subunit mutations in nonsyndromic mental retardation - PubMed (original) (raw)
doi: 10.1016/j.ajhg.2008.03.021. Epub 2008 May 1.
François Foulquier, Patrick S Tarpey, Willy Morelle, Sarah Boissel, Jon Teague, Sarah Edkins, P Andrew Futreal, Michael R Stratton, Gillian Turner, Gert Matthijs, Jozef Gecz, Arnold Munnich, Laurence Colleaux
Affiliations
- PMID: 18455129
- PMCID: PMC2427205
- DOI: 10.1016/j.ajhg.2008.03.021
Oligosaccharyltransferase-subunit mutations in nonsyndromic mental retardation
Florence Molinari et al. Am J Hum Genet. 2008 May.
Abstract
Mental retardation (MR) is the most frequent handicap among children and young adults. Although a large proportion of X-linked MR genes have been identified, only four genes responsible for autosomal-recessive nonsyndromic MR (AR-NSMR) have been described so far. Here, we report on two genes involved in autosomal-recessive and X-linked NSMR. First, autozygosity mapping in two sibs born to first-cousin French parents led to the identification of a region on 8p22-p23.1. This interval encompasses the gene N33/TUSC3 encoding one subunit of the oligosaccharyltransferase (OTase) complex, which catalyzes the transfer of an oligosaccharide chain on nascent proteins, the key step of N-glycosylation. Sequencing N33/TUSC3 identified a 1 bp insertion, c.787_788insC, resulting in a premature stop codon, p.N263fsX300, and leading to mRNA decay. Surprisingly, glycosylation analyses of patient fibroblasts showed normal N-glycan synthesis and transfer, suggesting that normal N-glycosylation observed in patient fibroblasts may be due to functional compensation. Subsequently, screening of the X-linked N33/TUSC3 paralog, the IAP gene, identified a missense mutation (c.932T-->G, p.V311G) in a family with X-linked NSMR. Recent studies of fucosylation and polysialic-acid modification of neuronal cell-adhesion glycoproteins have shown the critical role of glycosylation in synaptic plasticity. However, our data provide the first demonstration that a defect in N-glycosylation can result in NSMR. Together, our results demonstrate that fine regulation of OTase activity is essential for normal cognitive-function development, providing therefore further insights to understand the pathophysiological bases of MR.
Figures
Figure 1
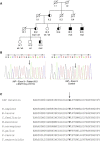
Genetic Analysis of Family 1 (A) Pedigree of family 1. The black symbols indicate the affected individuals. Autozygosity mapping was performed with Affymetrix GeneChip Human Mapping 10K for individuals IV.3, IV.4, and V.1 to V.4. Haplotypes are shown beneath each genotyped individual. Markers are from telomere to centromere, and their positions, based on the UCSC Genome browser, are indicated in bp. (B) Electropherograms of N33/TUSC3 exon 6 in an affected patient (left) and a control (right). DNA sequencing identified a one base-pair homozygous insertion c.787_788insC. (C) N33/TUSC3 mutation and phylogenetic analysis of N33/TUSC3 proteins in various species. Amino acids are in italics; bold and underlined indicate the frame shift caused by N33/TUSC3 mutation.
Figure 2
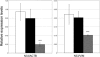
Quantitative PCR Analysis of N33/TUSC3 mRNA N33/TUSC3 expression in fibroblasts from two controls (black and white bars) and patient V.1 (gray bar). Data are normalized to Beta-actin (ACTB) or Vimentin (Vim). Means are given ± standard deviation (n = 4 to 8 independent RT-PCR). ∗∗∗, p < 0.001 as compared to controls, Student's t test.
Figure 3
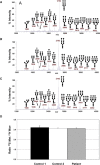
MALDI-TOF MS Analysis of Total Human Serum N-Glycome (A) Spectrum obtained from of a normal individual. (B and C) Spectra obtained from the two affected children of family 1, V.4 and V.1, respectively. Only the structures of the major N-glycans are given. A minor portion of the monofucosylated glycans carries fucose on an antenna rather than the core. Galactose (open circles), mannose (closed circles), GlcNAc (closed squares), fucose (open triangles), and NeuAc (closed diamonds) are shown. (D) Incorporation of [2-3H]mannose and [35S]methionine was determined after metabolic labeling of fibroblasts for 20, 40, and 60 min. Shown is the average ratio of [35S]methionine versus [2-3H]mannose incorporation into proteins of two independent experiments.
Figure 4
Genetic Analysis of Family 2 (A) Pedigree of family 2. The black symbols indicate individuals presenting severe MR, the white and black symbols indicate individuals presenting mild MR, and the white symbols indicate the nonaffected individuals. Carriers of mutated (mut) or wild-type (wt) alleles are indicated. (B) Electropherograms of IAP exon 9 in one affected boy of family 2 (left) and a control (right). DNA sequencing identified a missense mutation c.932T→G. (C) IAP mutation and phylogenetic analysis of IAP proteins in various species. The arrow and bold amino acids indicate the substitution caused by the mutation.
Figure 5
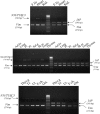
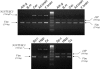
Expression Analyses of N33/TUSC3, IAP, and Vimentin Expression was asseyed by RT-PCR on adult and fetal total RNA isolated from various tissues (Clontech): fetal liver (F.liv), heart, kidney (Kid.), adult liver (Ad.liv), lung, placenta (Pl.), prostate (Pr.), salivary gland (Sal.g), skeletal muscle (Sk.m), testis, thymus, thyroid gland (Thyr.g), trachea (Tr.), uterus (Ut.), fetal chondrocytes (F.ch), and osteoblasts (Ost.). Amplicon length of N33/TUSC3, IAP, and Vimentin (Vim) are indicated in bp.
Figure 6
Expression Analyses of N33/TUSC3, IAP, and Vimentin (Vim) mRNAs in Adult and Fetal Brain Structures Expression was tested by RT-PCR on adult and fetal structures (Clontech): adrenal gland (Adr.g), bone marrow (B.m), cerebellum (Cer.), adult brain (Ad.brain), fetal brain (F.brain), spinal cord (Sp.c), hippocampus (Hipp.), and cortex (Cx). Amplicon length of N33/TUSC3, IAP, and Vimentin (Vim) are indicated in bp.
Similar articles
- A defect in the TUSC3 gene is associated with autosomal recessive mental retardation.
Garshasbi M, Hadavi V, Habibi H, Kahrizi K, Kariminejad R, Behjati F, Tzschach A, Najmabadi H, Ropers HH, Kuss AW. Garshasbi M, et al. Am J Hum Genet. 2008 May;82(5):1158-64. doi: 10.1016/j.ajhg.2008.03.018. Epub 2008 May 1. Am J Hum Genet. 2008. PMID: 18452889 Free PMC article. - Structural basis of substrate specificity of human oligosaccharyl transferase subunit N33/Tusc3 and its role in regulating protein N-glycosylation.
Mohorko E, Owen RL, Malojčić G, Brozzo MS, Aebi M, Glockshuber R. Mohorko E, et al. Structure. 2014 Apr 8;22(4):590-601. doi: 10.1016/j.str.2014.02.013. Epub 2014 Mar 27. Structure. 2014. PMID: 24685145 - Oligosaccharyltransferase: the central enzyme of N-linked protein glycosylation.
Mohorko E, Glockshuber R, Aebi M. Mohorko E, et al. J Inherit Metab Dis. 2011 Aug;34(4):869-78. doi: 10.1007/s10545-011-9337-1. Epub 2011 May 26. J Inherit Metab Dis. 2011. PMID: 21614585 Review. - Homozygous deletion in TUSC3 causing syndromic intellectual disability: a new patient.
Loddo S, Parisi V, Doccini V, Filippi T, Bernardini L, Brovedani P, Ricci F, Novelli A, Battaglia A. Loddo S, et al. Am J Med Genet A. 2013 Aug;161A(8):2084-7. doi: 10.1002/ajmg.a.36028. Epub 2013 Jul 4. Am J Med Genet A. 2013. PMID: 23825019 - TUSC3: a novel tumour suppressor gene and its functional implications.
Yu X, Zhai C, Fan Y, Zhang J, Liang N, Liu F, Cao L, Wang J, Du J. Yu X, et al. J Cell Mol Med. 2017 Sep;21(9):1711-1718. doi: 10.1111/jcmm.13128. Epub 2017 Mar 8. J Cell Mol Med. 2017. PMID: 28272772 Free PMC article. Review.
Cited by
- Community-based recruitment and exome sequencing indicates high diagnostic yield in adults with intellectual disability.
Sabo A, Murdock D, Dugan S, Meng Q, Gingras MC, Hu J, Muzny D, Gibbs R. Sabo A, et al. Mol Genet Genomic Med. 2020 Oct;8(10):e1439. doi: 10.1002/mgg3.1439. Epub 2020 Aug 7. Mol Genet Genomic Med. 2020. PMID: 32767738 Free PMC article. - Approaches to homozygosity mapping and exome sequencing for the identification of novel types of CDG.
Matthijs G, Rymen D, Millón MB, Souche E, Race V. Matthijs G, et al. Glycoconj J. 2013 Jan;30(1):67-76. doi: 10.1007/s10719-012-9445-7. Epub 2012 Sep 15. Glycoconj J. 2013. PMID: 22983704 Review. - Comprehensive Interactome Analysis Reveals that STT3B Is Required for N-Glycosylation of Lassa Virus Glycoprotein.
Zhu S, Wan W, Zhang Y, Shang W, Pan X, Zhang LK, Xiao G. Zhu S, et al. J Virol. 2019 Nov 13;93(23):e01443-19. doi: 10.1128/JVI.01443-19. Print 2019 Dec 1. J Virol. 2019. PMID: 31511384 Free PMC article. - SOBP is mutated in syndromic and nonsyndromic intellectual disability and is highly expressed in the brain limbic system.
Birk E, Har-Zahav A, Manzini CM, Pasmanik-Chor M, Kornreich L, Walsh CA, Noben-Trauth K, Albin A, Simon AJ, Colleaux L, Morad Y, Rainshtein L, Tischfield DJ, Wang P, Magal N, Maya I, Shoshani N, Rechavi G, Gothelf D, Maydan G, Shohat M, Basel-Vanagaite L. Birk E, et al. Am J Hum Genet. 2010 Nov 12;87(5):694-700. doi: 10.1016/j.ajhg.2010.10.005. Epub 2010 Oct 28. Am J Hum Genet. 2010. PMID: 21035105 Free PMC article. - Glycoprotein folding and quality-control mechanisms in protein-folding diseases.
Ferris SP, Kodali VK, Kaufman RJ. Ferris SP, et al. Dis Model Mech. 2014 Mar;7(3):331-41. doi: 10.1242/dmm.014589. Dis Model Mech. 2014. PMID: 24609034 Free PMC article. Review.
References
- American Psychiatric Association . 4th revised edition, DSM-IV-R. APA; Washington, D.C.: 1995. Diagnostic and Statistical Manual of Mental Disorders.
- Molinari F., Rio M., Meskenaite V., Encha-Razavi F., Auge J., Bacq D., Briault S., Vekemans M., Munnich A., Attie-Bitach T. Truncating neurotrypsin mutation in autosomal recessive nonsyndromic mental retardation. Science. 2002;298:1779–1781. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials