GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy - PubMed (original) (raw)
GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy
Yu Xue et al. Mol Cell Proteomics. 2008 Sep.
Abstract
Identification of protein phosphorylation sites with their cognate protein kinases (PKs) is a key step to delineate molecular dynamics and plasticity underlying a variety of cellular processes. Although nearly 10 kinase-specific prediction programs have been developed, numerous PKs have been casually classified into subgroups without a standard rule. For large scale predictions, the false positive rate has also never been addressed. In this work, we adopted a well established rule to classify PKs into a hierarchical structure with four levels, including group, family, subfamily, and single PK. In addition, we developed a simple approach to estimate the theoretically maximal false positive rates. The on-line service and local packages of the GPS (Group-based Prediction System) 2.0 were implemented in Java with the modified version of the Group-based Phosphorylation Scoring algorithm. As the first stand alone software for predicting phosphorylation, GPS 2.0 can predict kinase-specific phosphorylation sites for 408 human PKs in hierarchy. A large scale prediction of more than 13,000 mammalian phosphorylation sites by GPS 2.0 was exhibited with great performance and remarkable accuracy. Using Aurora-B as an example, we also conducted a proteome-wide search and provided systematic prediction of Aurora-B-specific substrates including protein-protein interaction information. Thus, the GPS 2.0 is a useful tool for predicting protein phosphorylation sites and their cognate kinases and is freely available on line.
Figures
Fig. 1.
The training data could be reused several times and included in different PK clusters based on their cognate PKs information.
Fig. 2.
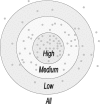
The basic idea of the Group-based Phosphorylation Scoring algorithm. The gray dots represent the positive sites. The nearer distances indicate higher similarity scores between two sites. Given a putative PSP(7, 7) peptide, we can calculate its score. Then we can judge whether the given site is a potentially real phosphorylation site under different thresholds.
Fig. 3.
A simple method of matrix mutation.
Fig. 4.
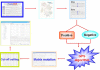
The process of construction of GPS 2.0 software. The training data were taken from the Phospho.ELM 6.0 database. All sites with kinase information were retained. Then these verified sites with their kinases were separated into a hierarchical structure with four levels, including group, family, subfamily, and single PK. The modified version of Group-based Phosphorylation Scoring algorithm was used. The matrix mutation was used to improve the robustness of the prediction system. Then we set the high, medium, and low thresholds based on the calculated FPR for each PK cluster. Finally GPS 2.0 was implemented in Java as the first stand alone software for computational phosphorylation.
Fig. 5.
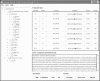
The screen snapshot of GPS 2.0 software. As an example, the protein sequence of rat Spinophilin was adopted. And the prediction results of PKA-specific sites with medium threshold are shown. DMPK, myotonic dystrophy protein kinase; PKC, protein kinase C; PKG, protein kinase G; RSK, ribosomal S6 kinase; SGK, serum- and glucocorticoid-regulated protein kinase; TKL, tyrosine kinase-like.
Fig. 6.
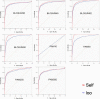
Comparison of various scoring matrices. Self, self-consistency. The BLOSUM62 matrix was adopted to balance the prediction performance and robustness of GPS 2.0.
Fig. 7.
Prediction performances before and after matrix mutations. For instance, we randomly chose 12 PK clusters to compare the performances. Usually the leave-one-out validations will be improved significantly. But the self-consistencies were only enhanced moderately. Thus, the process of matrix mutation improved both performance and robustness of GPS 2.0. MM, matrix mutation; Self, self-consistency; PKC, protein kinase C; RSK, ribosomal S6 kinase; MAPKAPK, MAPK-activated protein kinase.
Similar articles
- GPS 5.0: An Update on the Prediction of Kinase-specific Phosphorylation Sites in Proteins.
Wang C, Xu H, Lin S, Deng W, Zhou J, Zhang Y, Shi Y, Peng D, Xue Y. Wang C, et al. Genomics Proteomics Bioinformatics. 2020 Feb;18(1):72-80. doi: 10.1016/j.gpb.2020.01.001. Epub 2020 Mar 19. Genomics Proteomics Bioinformatics. 2020. PMID: 32200042 Free PMC article. - Proteome-wide prediction of PKA phosphorylation sites in eukaryotic kingdom.
Gao X, Jin C, Ren J, Yao X, Xue Y. Gao X, et al. Genomics. 2008 Dec;92(6):457-63. doi: 10.1016/j.ygeno.2008.08.013. Epub 2008 Oct 10. Genomics. 2008. PMID: 18817865 - GPS: a novel group-based phosphorylation predicting and scoring method.
Zhou FF, Xue Y, Chen GL, Yao X. Zhou FF, et al. Biochem Biophys Res Commun. 2004 Dec 24;325(4):1443-8. doi: 10.1016/j.bbrc.2004.11.001. Biochem Biophys Res Commun. 2004. PMID: 15555589 - The Aurora kinases: role in cell transformation and tumorigenesis.
Katayama H, Brinkley WR, Sen S. Katayama H, et al. Cancer Metastasis Rev. 2003 Dec;22(4):451-64. doi: 10.1023/a:1023789416385. Cancer Metastasis Rev. 2003. PMID: 12884918 Review. - PDK2: the missing piece in the receptor tyrosine kinase signaling pathway puzzle.
Dong LQ, Liu F. Dong LQ, et al. Am J Physiol Endocrinol Metab. 2005 Aug;289(2):E187-96. doi: 10.1152/ajpendo.00011.2005. Am J Physiol Endocrinol Metab. 2005. PMID: 16014356 Review.
Cited by
- GBMPhos: A Gating Mechanism and Bi-GRU-Based Method for Identifying Phosphorylation Sites of SARS-CoV-2 Infection.
Huang G, Xiao R, Chen W, Dai Q. Huang G, et al. Biology (Basel). 2024 Oct 6;13(10):798. doi: 10.3390/biology13100798. Biology (Basel). 2024. PMID: 39452107 Free PMC article. - RF-Phos: A Novel General Phosphorylation Site Prediction Tool Based on Random Forest.
Ismail HD, Jones A, Kim JH, Newman RH, Kc DB. Ismail HD, et al. Biomed Res Int. 2016;2016:3281590. doi: 10.1155/2016/3281590. Epub 2016 Mar 15. Biomed Res Int. 2016. PMID: 27066500 Free PMC article. - Attenphos: General Phosphorylation Site Prediction Model Based on Attention Mechanism.
Song T, Yang Q, Qu P, Qiao L, Wang X. Song T, et al. Int J Mol Sci. 2024 Jan 26;25(3):1526. doi: 10.3390/ijms25031526. Int J Mol Sci. 2024. PMID: 38338804 Free PMC article. - PhosTryp: a phosphorylation site predictor specific for parasitic protozoa of the family trypanosomatidae.
Palmeri A, Gherardini PF, Tsigankov P, Ausiello G, Späth GF, Zilberstein D, Helmer-Citterich M. Palmeri A, et al. BMC Genomics. 2011 Dec 19;12:614. doi: 10.1186/1471-2164-12-614. BMC Genomics. 2011. PMID: 22182631 Free PMC article. - Phosphorylation of mouse melanopsin by protein kinase A.
Blasic JR Jr, Brown RL, Robinson PR. Blasic JR Jr, et al. PLoS One. 2012;7(9):e45387. doi: 10.1371/journal.pone.0045387. Epub 2012 Sep 26. PLoS One. 2012. PMID: 23049792 Free PMC article.
References
- Zhou, F. F., Xue, Y., Yao, X., and Xu, Y. ( 2006) A general user interface for prediction servers of proteins’ post-translational modification sites. Nat. Protocol. 1, 1318–1321 - PubMed
- Blom, N., Gammeltoft, S., and Brunak, S. ( 1999) Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294, 1351–1362 - PubMed
- Ingrell, C. R., Miller, M. L., Jensen, O. N., and Blom, N. ( 2007) NetPhosYeast: prediction of protein phosphorylation sites in yeast. Bioinformatics 23, 895–897 - PubMed
- Tang, Y. R., Chen, Y. Z., Canchaya, C. A., and Zhang, Z. ( 2007) GANNPhos: a new phosphorylation site predictor based on a genetic algorithm integrated neural network. Protein Eng. Des. Sel. 20, 405–412 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous