Regulation of microtubule assembly and organization in mitosis by the AAA+ ATPase Pontin - PubMed (original) (raw)
Regulation of microtubule assembly and organization in mitosis by the AAA+ ATPase Pontin
Daniel Ducat et al. Mol Biol Cell. 2008 Jul.
Abstract
To identify novel proteins important for microtubule assembly in mitosis, we have used a centrosome-based complementation assay to enrich for proteins with mitotic functions. An RNA interference (RNAi)-based screen of these proteins allowed us to uncover 13 novel mitotic regulators. We carried out in-depth analyses of one of these proteins, Pontin, which is known to have several functions in interphase, including chromatin remodeling, DNA repair, and transcription. We show that reduction of Pontin by RNAi resulted in defects in spindle assembly in Drosophila S2 cells and in several mammalian tissue culture cell lines. Further characterization of Pontin in Xenopus egg extracts demonstrates that Pontin interacts with the gamma tubulin ring complex (gamma-TuRC). Because depletion of Pontin leads to defects in the assembly and organization of microtubule arrays in egg extracts, our studies suggest that Pontin has a mitosis-specific function in regulating microtubule assembly.
Figures
Figure 1.
Biochemical fractionation and proteomic analysis of centrosome-complementing activity. (A) Schematic for the biochemical enrichment of the centrosome-complementing activity. Crude extract made from Drosophila early embryos were first precipitated using 3% PEG, and the pellet was resuspended in buffer and fractionated on a sucrose gradient. The fractions with centrosome-complementing activity were pooled and subjected to Mono S chromatography for further purification of proteins that restore microtubule nucleation capacity to salt-stripped centrosomes. The crude extracts and certain fractions from each purification step above restored the ability of the KI-stripped Drosophila centrosomes to nucleate microtubules from purified tubulin (see Kawaguchi and Zheng, 2004; see Figure 4 for fraction analysis). (B) Identification of proteins present in the active fraction after Mono S chromatography by mass spectrometry. In total, 365 proteins were identified and grouped by their known or predicted functions (see Supplemental Table 1 for more details). (C) Representative proteins within the centrosome-complementing fraction that are known to associate with centrosomes or were identified as mitotic regulators by a recent whole-genome screen (Goshima et al., 2007).
Figure 2.
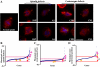
Effects of Pontin reduction by RNAi in mitotic S2 cells. (A) Examples of spindle and centrosome defects caused by Pontin depletion. S2 cells treated with Pontin RNAi were fixed and stained by DAPI (blue) and antibodies against α-tubulin (red) or CP309 (marking the centrosomes green). Shown are cells exhibiting spindles with disorganized or splayed poles (SSP), elongated and/or dim bipolar spindles (in the abnormal spindle, AS, category), misplaced centrosomes (CPD) relative to spindle poles, and/or low centrosome number (CNL). (B–D) Graphs of PSs. Pontin RNAi resulted in consistent (3 independent double-blind RNAi experiments; pink arrows) spindle defects (B), low centrosome number (C), and centrosome positioning defects (D) above the 95% CI (red lines). No consistent mitotic defects were seen in Reptin RNAi-treated cells (two independent double-blind RNAi experiments; yellow arrows). Images were acquired on a Leica SP5 confocal microscope at 1024 × 1024 resolution with two-line averaging. Bar, 5 μm.
Figure 3.
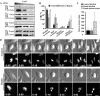
Effects of reduction of Pontin and Reptin on mammalian cell division and survival. (A) Reduction of Pontin and Reptin in HeLa cells by siRNA. Lysates made from HeLa cells treated for 72 h with control siRNA or siRNAs targeting Pontin and Reptin were fractionated by SDS-polyacrylamide gel electrophoresis in decreasing amounts (16, 12, or 8 μl) and immunoblotted for the indicated proteins. RNAi treatment resulted in ∼65–75% or 85–95% reduction of Pontin or Reptin, respectively, compared with controls. (B–D) Analyses of Pontin and Reptin RNAi by live imaging. siRNA-treated HeLa cells expressing Histone H2B-GFP were imaged for 72 h beginning 24 h after transfection. Graph in B shows percentages of cells completing division, dying in mitosis, or dying in interphase after Pontin, Reptin, or control siRNA treatment. Error bars, standard deviations calculated from at least four independent experiments where at least 30 mitotic events were analyzed in each experiment. Whereas both Pontin and Reptin siRNA-treatment caused interphase cell death, only Pontin siRNA caused mitotic cell death. Graph in C shows that Pontin RNAi-treated cells that died in mitosis first underwent mitotic arrest and then committed death, whereas the cells that divided successfully had similar mitotic timing as control cells. Error bars, standard deviations calculated from three independent experiments where at least 50 mitotic events were analyzed in each experiment. The mitotic timing was calculated from nuclear envelope breakdown to the onset of anaphase or cell death. (D) Still images of control or Pontin siRNA-treated HeLa cells entering mitosis. Brightfield and fluorescence images captured at indicated time frame (hours:minutes) are shown. Time set to “0:00” upon nuclear envelope breakdown. Note that Pontin-depleted cells arrest in prometaphase with partially aligned chromosomes (arrows) before undergoing mitotic death, judged by cell membrane blebbing and DNA compaction (arrowheads). Bar, 10 μm.
Figure 4.
Effects of Pontin reduction on mitotic spindle assembly in mammalian tissue culture cells. (A) Spindle pole localization of Pontin. U2OS cells treated with either control or Pontin siRNA were fixed in methanol and stained by DAPI (blue), antibodies to α-tubulin (red), and peptide antibodies to Pontin (green). Note that the spindle pole staining of Pontin (top) diminished after treatment with Pontin siRNA (bottom). Images were acquired on a Nikon E800. (B) Displacement of γ-tubulin from mitotic spindles after Pontin depletion. γ-Tubulin staining (green) in Pontin-depleted cells was reduced both at the mitotic spindle poles and along the spindles with a corresponding increase in cytoplasmic staining (bottom), relative to control-treated cells (top). Images shown are maximum projections of z-stacks acquired on a Yokogawa CSU10 spinning disk confocal microscope. (C) Spindle defects in Pontin-depleted cells. Spindle pole fragmentation and multipolar spindles were the most frequent defects observed after Pontin depletion, as viewed by pericentrin staining (green). Images were acquired on a Nikon E800. (D) Quantification of abnormal and multipolar spindles in U2OS cells 48–72 h after control, Pontin (3 oligos), or Reptin (2 oligos) siRNA treatment. Similar increases in spindle defects after Pontin reduction were observed in HeLa and MCF10-A cells (data not shown). (E) Quantification of the ratio of γ-tubulin intensity at bipolar spindle poles to the intensity of a representative area of the cytoplasm (see Materials and Methods). Error bars in D and E represent SD from at least three independent experiments where >50 mitotic cells or >30 mitotic poles were counted or measured in each experiment, respectively. Bars, 10 μm.
Figure 5.
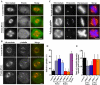
Interactions among Pontin, Reptin, and γTuRC in Xenopus egg extracts. (A) Specificity of antibodies generated against a peptide of Xenopus Pontin (Pep-Pontin) and full-length Pontin (FL-Pontin) and Reptin (FL-Reptin) by Western blotting analysis using Xenopus egg extracts. (B) Localization of Pontin on spindles. RanGTP-induced spindles in Xenopus egg extracts were fixed in methanol and stained with anti-α-tubulin (red) and peptide anti-Pontin (green) antibodies. Pontin localized to spindle poles and lightly along spindle microtubules. (C) Interaction between Pontin and γTuRC in egg extracts. Control IgG, Pontin, or γ-tubulin antibodies were used in immunoprecipitation reactions. Pontin and γ-tubulin immunoprecipitated each other reciprocally. Each antibody also coprecipitated Xgrip109 and Xgrip210, other components of γTuRC. (D) Comigration of Pontin and Reptin in Xenopus egg extracts on sucrose gradients. Protein standards with S values of 4.3 S (bovine serum albumin), 7.3 S (rabbit muscle aldolase), 11.3 S (bovine liver catalase), and 19.4 S (porcine thyroglobulin) were run on identical gradients. Peaks corresponding to these protein standards are indicated (arrowheads). (E) Copurification of Pontin and Reptin with γTuRC. Purified γTuRC complex (Coomassie Blue stained; see Materials and Methods) contained substoichiometric amounts of both Pontin and Reptin as revealed by Western blotting analysis. (F) Reptin antibodies fail to immunoprecipitate the Reptin associated with γTuRC in Xenopus egg extracts. Xenopus egg extracts were either mock-depleted using unimmunized rabbit IgG or depleted of Reptin (and associated Pontin) using Reptin-specific antibodies. Different amounts of egg extracts (0.75, 0.25, and 0.1 μl) were analyzed by Western blotting (top). Approximately 85–95% of the total pool of Reptin (and Pontin; data not shown) was depleted. After IgG or Reptin depletion, γTuRC was immunoprecipitated from egg extracts using γ-tubulin antibodies. The immunoprecipitates were analyzed by loading decreasing amounts of sample (12, 8, and 4 μl) on the gel. The same amount of Pontin and Reptin remained associated with the γTuRC in Reptin depleted egg extracts as compared with mock-depleted ones (bottom), demonstrating that Reptin antibodies failed to recognize the γTuRC-associated Reptin in the egg extracts. Bar, 10 μm.
Figure 6.
Requirement for Pontin in microtubule assembly in Xenopus egg extracts. (A) Western blotting analyses of egg extracts that were mock depleted (−IgG), depleted with Pontin antibodies (−Pontin), depleted of Pontin followed by γTuRC addition (−Pontin+γTuRC), or depleted of Pontin followed by addition of γTuRC, Pontin, and Reptin (−Pontin+γTuRC+Pont/Rept). Immunodepletion of Pontin from the egg extracts codepleted both γTuRC components and Reptin. Endogenous levels of γTuRC, Pontin, and Reptin were restored by addition of purified proteins. Effects of protein depletion on microtubule assembly are assayed in C. (B) Examples of AurA beads associated with a spindle-like structure (arrow), bright microtubule asters (filled arrowheads), faint microtubule asters (open arrowheads), or no microtubules. (C) Quantification of AurA-beads associated with spindles or bright microtubule asters in egg extracts treated as in A. Microtubule structures in controls were normalized to 100%. Depletion of Pontin abrogated the capacity of AurA beads to stimulate assembly of spindles and microtubule asters as compared with mock depletion. Addition of purified γTuRC resulted in partial rescue. Full rescue of microtubule nucleation was only observed upon addition of purified Pontin and Reptin along with γTuRC. Error bars, SD from >10 independent experiments. The p values were calculated using a two-tailed Student's t test. Bar, 10 μm.
Similar articles
- CHD4 is a RanGTP-dependent MAP that stabilizes microtubules and regulates bipolar spindle formation.
Yokoyama H, Nakos K, Santarella-Mellwig R, Rybina S, Krijgsveld J, Koffa MD, Mattaj IW. Yokoyama H, et al. Curr Biol. 2013 Dec 16;23(24):2443-51. doi: 10.1016/j.cub.2013.09.062. Epub 2013 Nov 21. Curr Biol. 2013. PMID: 24268414 - The nucleoporin MEL-28 promotes RanGTP-dependent γ-tubulin recruitment and microtubule nucleation in mitotic spindle formation.
Yokoyama H, Koch B, Walczak R, Ciray-Duygu F, González-Sánchez JC, Devos DP, Mattaj IW, Gruss OJ. Yokoyama H, et al. Nat Commun. 2014;5:3270. doi: 10.1038/ncomms4270. Nat Commun. 2014. PMID: 24509916 - Relocalization of human chromatin remodeling cofactor TIP48 in mitosis.
Sigala B, Edwards M, Puri T, Tsaneva IR. Sigala B, et al. Exp Cell Res. 2005 Nov 1;310(2):357-69. doi: 10.1016/j.yexcr.2005.07.030. Epub 2005 Sep 12. Exp Cell Res. 2005. PMID: 16157330 - RVB1/RVB2: running rings around molecular biology.
Jha S, Dutta A. Jha S, et al. Mol Cell. 2009 Jun 12;34(5):521-33. doi: 10.1016/j.molcel.2009.05.016. Mol Cell. 2009. PMID: 19524533 Free PMC article. Review. - Chaperone-like activity of the AAA+ proteins Rvb1 and Rvb2 in the assembly of various complexes.
Nano N, Houry WA. Nano N, et al. Philos Trans R Soc Lond B Biol Sci. 2013 Mar 25;368(1617):20110399. doi: 10.1098/rstb.2011.0399. Print 2013 May 5. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23530256 Free PMC article. Review.
Cited by
- Chromosome Missegregation Associated with RUVBL1 Deficiency.
Gentili C, Castor D, Kaden S, Lauterbach D, Gysi M, Steigemann P, Gerlich DW, Jiricny J, Ferrari S. Gentili C, et al. PLoS One. 2015 Jul 22;10(7):e0133576. doi: 10.1371/journal.pone.0133576. eCollection 2015. PLoS One. 2015. PMID: 26201077 Free PMC article. - The multi-faceted roles of R2TP complex span across regulation of gene expression, translation, and protein functional assembly.
Luthuli SD, Shonhai A. Luthuli SD, et al. Biophys Rev. 2023 Sep 12;15(6):1951-1965. doi: 10.1007/s12551-023-01127-9. eCollection 2023 Dec. Biophys Rev. 2023. PMID: 38192347 Free PMC article. Review. - Knockdown of DOM/Tip60 Complex Subunits Impairs Male Meiosis of Drosophila melanogaster.
Prozzillo Y, Fattorini G, Ferreri D, Leo M, Dimitri P, Messina G. Prozzillo Y, et al. Cells. 2023 May 9;12(10):1348. doi: 10.3390/cells12101348. Cells. 2023. PMID: 37408183 Free PMC article. - Specific threonine-4 phosphorylation and function of RNA polymerase II CTD during M phase progression.
Hintermair C, Voß K, Forné I, Heidemann M, Flatley A, Kremmer E, Imhof A, Eick D. Hintermair C, et al. Sci Rep. 2016 Jun 6;6:27401. doi: 10.1038/srep27401. Sci Rep. 2016. PMID: 27264542 Free PMC article. - MTBP and MYC: A Dynamic Duo in Proliferation, Cancer, and Aging.
Grieb BC, Eischen CM. Grieb BC, et al. Biology (Basel). 2022 Jun 8;11(6):881. doi: 10.3390/biology11060881. Biology (Basel). 2022. PMID: 35741402 Free PMC article. Review.
References
- Bettencourt-Dias M., et al. Genome-wide survey of protein kinases required for cell cycle progression. Nature. 2004;432:980–987. - PubMed
- Blagosklonny M. V. Mitotic arrest and cell fate: why and how mitotic inhibition of transcription drives mutually exclusive events. Cell Cycle. 2007;6:70–74. - PubMed
- Blower M. D., Nachury M., Heald R., Weis K. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 2005;121:223–234. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 MH067880/MH/NIMH NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 GM056312/GM/NIGMS NIH HHS/United States
- MH-067880/MH/NIMH NIH HHS/United States
- P41 RR011823/RR/NCRR NIH HHS/United States
- GM-56312/GM/NIGMS NIH HHS/United States
- P41 RR-011823/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous