Resolution among major placental mammal interordinal relationships with genome data imply that speciation influenced their earliest radiations - PubMed (original) (raw)
Resolution among major placental mammal interordinal relationships with genome data imply that speciation influenced their earliest radiations
Björn M Hallström et al. BMC Evol Biol. 2008.
Abstract
Background: A number of the deeper divergences in the placental mammal tree are still inconclusively resolved despite extensive phylogenomic analyses. A recent analysis of 200 kbp of protein coding sequences yielded only limited support for the relationships among Laurasiatheria (cow, dog, bat and shrew), probably because the divergences occurred only within a few million years from each other. It is generally expected that increasing the amount of data and improving the taxon sampling enhance the resolution of narrow divergences. Therefore these and other difficult splits were examined by phylogenomic analysis of the hitherto largest sequence alignment. The increasingly complete genome data of placental mammals also allowed developing a novel and stringent data search method.
Results: The rigorous data handling, recursive BLAST, successfully removed the sequences from gene families, including those from well-known families hemoglobin, olfactory, myosin and HOX genes, thus avoiding alignment of possibly paralogous sequences. The current phylogenomic analysis of 3,012 genes (2,844,615 nucleotides) from a total of 22 species yielded statistically significant support for most relationships. While some major clades were confirmed using genomic sequence data, the placement of the treeshrew, bat and the relationship between Boreoeutheria, Xenarthra and Afrotheria remained problematic to resolve despite the size of the alignment. Phylogenomic analysis of divergence times dated the basal placental mammal splits at 95-100 million years ago. Many of the following divergences occurred only a few (2-4) million years later. Relationships with narrow divergence time intervals received unexpectedly limited support even from the phylogenomic analyses.
Conclusion: The narrow temporal window within which some placental divergences took place suggests that inconsistencies and limited resolution of the mammalian tree may have their natural explanation in speciation processes such as lineage sorting, introgression from species hybridization or hybrid speciation. These processes obscure phylogenetic analysis, making some parts of the tree difficult to resolve even with genome data.
Figures
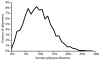
Figure 1
Distance distribution of the human-platypus aa sequence alignment.
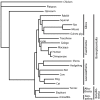
Figure 2
ML tree on NT12 analysis and ML bootstrap support values (nt/aa) for branches that do not receive maximum support.
Figure 3
Partial trees illustrating the alternative topologies that were statistically evaluated to determine the phylogenetic position of the tree shrew (1–3), the relationships within Rodentia (4–6), the position of the bat (7–11), and the Xenarthra, Afrotheria and Boreoplacentalia relationships (12–14).
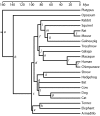
Figure 4
Chronogram showing the estimated divergence times. The figure is based on divergence times estimated from nt sequences using TF. Divergences are labeled with letters (a-t) and the exact dates and the values estimated by other methods and sequence data are shown in Table 3.
Similar articles
- Mammalian evolution may not be strictly bifurcating.
Hallström BM, Janke A. Hallström BM, et al. Mol Biol Evol. 2010 Dec;27(12):2804-16. doi: 10.1093/molbev/msq166. Epub 2010 Jun 29. Mol Biol Evol. 2010. PMID: 20591845 Free PMC article. - Phylogenomic data analyses provide evidence that Xenarthra and Afrotheria are sister groups.
Hallström BM, Kullberg M, Nilsson MA, Janke A. Hallström BM, et al. Mol Biol Evol. 2007 Sep;24(9):2059-68. doi: 10.1093/molbev/msm136. Epub 2007 Jul 13. Mol Biol Evol. 2007. PMID: 17630282 - Phylogenomic analysis resolves the interordinal relationships and rapid diversification of the laurasiatherian mammals.
Zhou X, Xu S, Xu J, Chen B, Zhou K, Yang G. Zhou X, et al. Syst Biol. 2012 Jan;61(1):150-64. doi: 10.1093/sysbio/syr089. Epub 2011 Sep 7. Syst Biol. 2012. PMID: 21900649 Free PMC article. - Mammal madness: is the mammal tree of life not yet resolved?
Foley NM, Springer MS, Teeling EC. Foley NM, et al. Philos Trans R Soc Lond B Biol Sci. 2016 Jul 19;371(1699):20150140. doi: 10.1098/rstb.2015.0140. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 27325836 Free PMC article. Review. - The chromosomes of Afrotheria and their bearing on mammalian genome evolution.
Svartman M, Stanyon R. Svartman M, et al. Cytogenet Genome Res. 2012;137(2-4):144-53. doi: 10.1159/000341387. Epub 2012 Aug 3. Cytogenet Genome Res. 2012. PMID: 22868637 Review.
Cited by
- How to Make a Dolphin: Molecular Signature of Positive Selection in Cetacean Genome.
Nery MF, González DJ, Opazo JC. Nery MF, et al. PLoS One. 2013 Jun 19;8(6):e65491. doi: 10.1371/journal.pone.0065491. Print 2013. PLoS One. 2013. PMID: 23840335 Free PMC article. - Mammalian evolution may not be strictly bifurcating.
Hallström BM, Janke A. Hallström BM, et al. Mol Biol Evol. 2010 Dec;27(12):2804-16. doi: 10.1093/molbev/msq166. Epub 2010 Jun 29. Mol Biol Evol. 2010. PMID: 20591845 Free PMC article. - Accelerated evolutionary rate of the myoglobin gene in long-diving whales.
Nery MF, Arroyo JI, Opazo JC. Nery MF, et al. J Mol Evol. 2013 Jun;76(6):380-7. doi: 10.1007/s00239-013-9572-1. Epub 2013 Jul 16. J Mol Evol. 2013. PMID: 23857304 - A connection to the past: Monodelphis domestica provides insight into the organization and connectivity of the brains of early mammals.
Dooley JC, Franca JG, Seelke AM, Cooke DF, Krubitzer LA. Dooley JC, et al. J Comp Neurol. 2013 Dec 1;521(17):3877-97. doi: 10.1002/cne.23383. J Comp Neurol. 2013. PMID: 23784751 Free PMC article. - Evolutionary Models for the Diversification of Placental Mammals Across the KPg Boundary.
Springer MS, Foley NM, Brady PL, Gatesy J, Murphy WJ. Springer MS, et al. Front Genet. 2019 Nov 29;10:1241. doi: 10.3389/fgene.2019.01241. eCollection 2019. Front Genet. 2019. PMID: 31850081 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials