Autoinhibition of human dicer by its internal helicase domain - PubMed (original) (raw)
Autoinhibition of human dicer by its internal helicase domain
Enbo Ma et al. J Mol Biol. 2008.
Abstract
Dicer, a member of the ribonuclease III family of enzymes, processes double-stranded RNA substrates into approximately 21- to 27-nt products that trigger sequence-directed gene silencing by RNA interference. Although the mechanism of RNA recognition and length-specific cleavage by Dicer has been established, the way in which dicing activity is regulated is unclear. Here, we show that the N-terminal domain of human Dicer, which is homologous to DExD/H-box helicases, substantially attenuates the rate of substrate cleavage. Deletion or mutation of this domain activates human Dicer in both single- and multiple-turnover assays. The catalytic efficiency (k(cat)/K(m)) of the deletion construct is increased by 65-fold over that exhibited by the intact enzyme. Kinetic analysis shows that this activation is almost entirely due to an enhancement in k(cat). Modest stimulation of catalysis by the full-length Dicer enzyme was observed in the presence of the TAR-RNA binding protein, which physically interacts with the DExD/H-box domain. These results suggest that the DExD/H-box domain likely disrupts the functionality of the Dicer active site until a structural rearrangement occurs, perhaps upon assembly with its molecular partners.
Figures
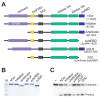
Figure 1. Domain structure and expression of human Dicer (hDcr)
A, Domain structure of hDcr variants; B, 10% SDS-PAGE analysis of recombinant hDcr proteins; C, Single time-point dsRNase activity assays (60 min., 37□, 60 nM protein, 2 nM 32P-labeled 37ab RNA); M, protein size marker.
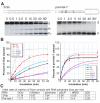
Figure 2. Single-turnover activity of hDcr proteins
A, Substrates and dicing reactions. Left panels, perfect-duplex dsRNA substrate, 37ab; right panels, hairpin pre-miRNA substrate, pre-hlet-7; asterisks (*) indicate 5′-end labeled with 32P. hDcr generates two products from RNA 37ab, 22-nt (P1) or 15-nt (P2), and one product (P) from pre-hlet-7. Relative rates of P1 and P2 production were the same for all hDcrs tested and were combined to give the total cleavage product for 37ab. B, Single-turnover reaction of hDcrs (60 nM) with 2 nM (3000 c.p.m.) duplex RNA 37ab (left panel) or pre-hlet-7 (right panel); values are the average from two independent experiments. Data were fit to the equation S_=(a-b)exp(−_k_obsd_t)+b, where S is the fraction of dsRNA cleaved at each time point, a is the fraction of dsRNA at the beginning of the reaction, b is the fraction of dsRNA at the reaction plateau (t-->∞), and _k_obsd is the observed rate constant; _k_obsd values (fmol/min) for the 37ab substrate were as follows: hDcr (wildtype), 0.32; hWalker, 0.8; Δhelicase (Δhel), 2; ΔDUF, 0.0034; ΔRBD, 0.2; 2DD, 0.46; _k_obsd values (fmol/min) for the pre-hlet-7 substrate were as follows: hDcr (wildtype), 3; hWalker, 2; Δhelicase, 2.2; ΔDUF, 2.6; ΔRBD, 0.74; 2DD, 0.74; C, summary of initial reaction rates of hDcr variants calculated for 20% substrate cleavage; S, RNA substrate; FL, wild-type hDicer; hW, hWalker mutant; dhel, Δhelicase mutant; dDUF, ΔDUF mutant; dRBD, ΔRBD mutant.
Figure 3. Duplex RNA/hDcr dissociation constants
Equilibrium filter binding assays for hDcr proteins complexed with 37ab RNA (A) or pre-hlet-7 RNA (B). Values are averages from two independent assays. C, summary of dissociation constants (Kd, nM).
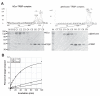
Figure 4. Steady-state kinetic analysis of wild-type and Δhelicase hDcr constructs
Plots of initial velocity versus substrate concentration: A, hDcr; and B, Δhelicase. C, summary of kinetic values.
Figure 5. Dicer-TRBP interaction and kinetics
A, size exclusion chromatography of the hDcr-TRBP complex formed by incubation of hDcr (molecular weight 219 kDa; 2.5 nmol) and excess TRBP (molecular weight 39 kDa; 9 nmol) in 20 μl dicing buffer for 60 min. on ice; chromatogram peak C, hDcr-TRBP complex; chromatogram peak T, TRBP; chromatogram peak hD, hDcr. SDS-PAGE gel analysis of fractions are shown below each chromatogram. B, multiple-turnover assay for 37ab cleavage using 100 nM dsRNA and 5 nM hDcr or chromatographically pure hDcr-TRBP; the data shown are representative of two independent experiments.
Similar articles
- Homodimeric structure and double-stranded RNA cleavage activity of the C-terminal RNase III domain of human dicer.
Takeshita D, Zenno S, Lee WC, Nagata K, Saigo K, Tanokura M. Takeshita D, et al. J Mol Biol. 2007 Nov 16;374(1):106-20. doi: 10.1016/j.jmb.2007.08.069. Epub 2007 Sep 8. J Mol Biol. 2007. PMID: 17920623 - DUF283 domain of Dicer proteins has a double-stranded RNA-binding fold.
Dlakić M. Dlakić M. Bioinformatics. 2006 Nov 15;22(22):2711-4. doi: 10.1093/bioinformatics/btl468. Epub 2006 Sep 5. Bioinformatics. 2006. PMID: 16954143 - Cryo-EM structures of human DICER dicing a pre-miRNA substrate.
Lee H, Roh SH. Lee H, et al. FEBS J. 2024 Jul;291(14):3072-3079. doi: 10.1111/febs.17048. Epub 2024 Jan 10. FEBS J. 2024. PMID: 38151772 Review. - Single processing center models for human Dicer and bacterial RNase III.
Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Zhang H, et al. Cell. 2004 Jul 9;118(1):57-68. doi: 10.1016/j.cell.2004.06.017. Cell. 2004. PMID: 15242644 - The mechanism of RNase III action: how dicer dices.
Ji X. Ji X. Curr Top Microbiol Immunol. 2008;320:99-116. doi: 10.1007/978-3-540-75157-1_5. Curr Top Microbiol Immunol. 2008. PMID: 18268841 Review.
Cited by
- CRISPR-Induced Expression of N-Terminally Truncated Dicer in Mouse Cells.
Malik R, Svoboda P. Malik R, et al. Genes (Basel). 2021 Apr 8;12(4):540. doi: 10.3390/genes12040540. Genes (Basel). 2021. PMID: 33918028 Free PMC article. - Untangling the roles of RNA helicases in antiviral innate immunity.
Baldaccini M, Pfeffer S. Baldaccini M, et al. PLoS Pathog. 2021 Dec 9;17(12):e1010072. doi: 10.1371/journal.ppat.1010072. eCollection 2021 Dec. PLoS Pathog. 2021. PMID: 34882751 Free PMC article. Review. - Slicing and dicing viruses: antiviral RNA interference in mammals.
Maillard PV, van der Veen AG, Poirier EZ, Reis e Sousa C. Maillard PV, et al. EMBO J. 2019 Apr 15;38(8):e100941. doi: 10.15252/embj.2018100941. Epub 2019 Mar 14. EMBO J. 2019. PMID: 30872283 Free PMC article. Review. - Crosstalk Between Mammalian Antiviral Pathways.
Watson SF, Knol LI, Witteveldt J, Macias S. Watson SF, et al. Noncoding RNA. 2019 Mar 22;5(1):29. doi: 10.3390/ncrna5010029. Noncoding RNA. 2019. PMID: 30909383 Free PMC article. Review. - Replication of many human viruses is refractory to inhibition by endogenous cellular microRNAs.
Bogerd HP, Skalsky RL, Kennedy EM, Furuse Y, Whisnant AW, Flores O, Schultz KL, Putnam N, Barrows NJ, Sherry B, Scholle F, Garcia-Blanco MA, Griffin DE, Cullen BR. Bogerd HP, et al. J Virol. 2014 Jul;88(14):8065-76. doi: 10.1128/JVI.00985-14. Epub 2014 May 7. J Virol. 2014. PMID: 24807715 Free PMC article.
References
- Carmell MA, Hannon GJ. RNase III enzymes and the initiation of gene silencing. Nature Struct. Mol. Biol. 2004;11:214–8. - PubMed
- Murchison EP, Hannon GJ. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Current Opinion Cell Biol. 2004;16:223–9. - PubMed
- MacRae IJ, Doudna JA. Ribonuclease revisited: structural insights into ribonuclease III family enzymes. Current Opinion Struct. Biol. 2007;17:138–45. - PubMed
- Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nature Struct. Mol. Biol. 2003;10:1026–32. - PubMed
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous