Recurrent gene fusions in prostate cancer - PubMed (original) (raw)
Review
Recurrent gene fusions in prostate cancer
Chandan Kumar-Sinha et al. Nat Rev Cancer. 2008 Jul.
Abstract
The discovery of recurrent gene fusions in a majority of prostate cancers has important clinical and biological implications in the study of common epithelial tumours. Gene fusion and chromosomal rearrangements were previously thought to be primarily the oncogenic mechanism of haematological malignancies and sarcomas. The prostate cancer gene fusions that have been identified thus far are characterized by 5' genomic regulatory elements, most commonly controlled by androgen, fused to members of the Ets family of transcription factors, leading to the overexpression of oncogenic transcription factors. Ets gene fusions probably define a distinct class of prostate cancer, and this might have a bearing on diagnosis, prognosis and rational therapeutic targeting.
Figures
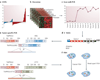
FIGURE 1. Cytological and Molecular Techniques used in Gene Fusion Studies
A. Cancer Outlier Profile Analysis (COPA) (described in the main text) was applied to microarray data available Oncomine (B.), and the top “outlier” genes formed our primary candidates to test for potential genomic rearrangements using cytogenetic and molecular techniques. C. The downstream workflow begins with quantitative real time PCR specific to exons from all parts of the gene (Exon-walk PCR), whereby a sharply divergent expression pattern from different parts of the gene suggests that a part of the gene may be split/ rearranged. D. Fusion partners of candidate split genes are identified by cloning the beginning and end of the candidate transcript using rapid amplification of cDNA ends (RACE). 5’ RACE is used to identify the 5’ fusion partner, and involves ligating an RNA adaptor sequence to the test RNA and carrying out RT-PCR using RNA adaptor specific forward primer and candidate gene specific reverse primer (located close to the split exon). The RACE PCR products are cloned and sequenced to confirm the identity of fusion genes. E. Fusion specific RT-PCR is used to confirm the presence of fusion transcript. F. Finally, the chromosomal rearrangements involved in the gene fusion are characterized by fluorescence in-situ hybridization (FISH) on interphase nuclei. FISH analysis involves hybridization of fluorescently labeled DNA probes corresponding to the candidate target sequences. Since it allows visualization of specific regions of the chromosome based on DNA sequence, it is the method of choice to investigate the genomic rearrangements like insertion, deletion, and translocation that can split or fuse genes. In the ‘split probe’ or ‘break apart’ assay strategy, two fluorescent probes (for example, green and red) are hybridized to sequences less than 1Mb apart, so that in normal interphase nucleus they appear co-localized. If a genetic rearrangement involves the region spanned by the two probes, the signals are separated and appear distinct (translocation), or a single color signal is seen and the other signal is lost (deletion). In the alternative ‘fusion probe’ strategy, two probes are localized far apart on the chromosome- or on two separate chromosomes. Two pairs of separate signals are seen in the normal diploid nucleus and in the event of a chromosomal rearrangement leading to gene fusion, the two signals are co-localized. Both these techniques have been used in the gene fusion studies, often in combination.
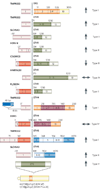
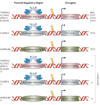
FIGURE 2. Anatomy of Gene fusions in Prostate Cancer
A cartoon representation of the gene fusions characterized in prostate cancers so far. The 5’ fusion partners are depicted on the left side and corresponding 3’ partners on the right. Light colors at the ends of the genes depict untranslated exons. The dark colored boxes depict coding exons. The numbers on the boxes identify the base positions of the exons. The arrows represent androgen responsiveness of the fusion genes, arrows pointing up (red) signify androgen mediated upregulation; arrows pointing down (green) represent androgen mediated down regulation of the corresponding gene; the horizontal arrows (blue) represent absence of androgen action on the fusion genes’ expression. TMPRSS2-ETS gene fusions have been grouped as Type I; other gene fusions which are androgen inducible have been grouped as Type II, androgen repressed fusion genes make up Type III, Androgen insensitive fusion genes, Type IV, and lastly, the novel situation in prostate cancer cell lines, with ETS genes rearranged to an androgen sensitive location (without the generation of classical gene fusions) has been classified as Type V.
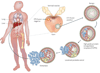
FIGURE 3. Genesis and Progression of Gene Fusions in Prostate Cancer
ETS gene fusions can first be observed in a subset of high grade prostatic intraepithelial neoplasia (HGPINs). Intriguingly, these foci are nearly invariably associated with intermingling foci of carcinoma, suggesting that ETS gene fusions may drive the progression to localized prostate cancer. Alternative lesions likely drive the development of ETS gene fusion negative HGPIN and localized carcinoma. While the same prostate may harbor both fusion positive and negative tumor foci, only a single foci metastasizes to distant organs, as different metastatic sites in the same patient are uniformly fusion positive or negative.
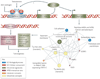
FIGURE 4. Role of Gene Fusions in Prostate Cancer
Androgen mediated upregulation of ETS genes (for example ERG or ETV1) leads to the activation of ETS target genes that upregulate various “Molecular Concepts” as identified by Molecular Concept Modeling (MCM), , and comprise genes involved in increased protein synthesis, cell adhesion and invasion including Urokinase plasminogen activator (PLAU) and several matrix metalloproteases. Preceding genetic lesions, including the loss of a tumor suppressor such as phosphatase and tensin homolog (PTEN), likely drive the development of PIN, while subsequent ETS gene mediated molecular alterations lead to the development of invasive prostate carcinogenesis.
FIGURE 5. Gamut of Gene Fusions in Prostate Cancer
Six major classes of pathognomic gene fusions in prostate cancer are envisioned based on the types of upstream regulatory partners and the nature of the fused genes. These include regulatory regions that are either androgen responsive or androgen repressed or androgen insensitive (housekeeping) fused to ETS family genes or other, yet to be discovered, non-ETS genes
Similar articles
- Bioinformatics approach leads to the discovery of the TMPRSS2:ETS gene fusion in prostate cancer.
Rubin MA, Chinnaiyan AM. Rubin MA, et al. Lab Invest. 2006 Nov;86(11):1099-102. doi: 10.1038/labinvest.3700477. Epub 2006 Sep 18. Lab Invest. 2006. PMID: 16983328 - Recurrent gene fusions in prostate cancer: their clinical implications and uses.
Hessels D, Schalken JA. Hessels D, et al. Curr Urol Rep. 2013 Jun;14(3):214-22. doi: 10.1007/s11934-013-0321-1. Curr Urol Rep. 2013. PMID: 23625457 Review. - The discovery and application of gene fusions in prostate cancer.
Morris DS, Tomlins SA, Montie JE, Chinnaiyan AM. Morris DS, et al. BJU Int. 2008 Aug;102(3):276-82. doi: 10.1111/j.1464-410X.2008.07665.x. Epub 2008 Apr 16. BJU Int. 2008. PMID: 18422767 Free PMC article. Review. - Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer.
Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, Yu J, Wang L, Montie JE, Rubin MA, Pienta KJ, Roulston D, Shah RB, Varambally S, Mehra R, Chinnaiyan AM. Tomlins SA, et al. Nature. 2007 Aug 2;448(7153):595-9. doi: 10.1038/nature06024. Nature. 2007. PMID: 17671502
Cited by
- BET bromodomain-mediated interaction between ERG and BRD4 promotes prostate cancer cell invasion.
Blee AM, Liu S, Wang L, Huang H. Blee AM, et al. Oncotarget. 2016 Jun 21;7(25):38319-38332. doi: 10.18632/oncotarget.9513. Oncotarget. 2016. PMID: 27223260 Free PMC article. - Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage.
Stork CT, Bocek M, Crossley MP, Sollier J, Sanz LA, Chédin F, Swigut T, Cimprich KA. Stork CT, et al. Elife. 2016 Aug 23;5:e17548. doi: 10.7554/eLife.17548. Elife. 2016. PMID: 27552054 Free PMC article. - Clinically relevant genetic characterization of prostate tumors: how close are we to the goal?
Tolkach Y, Imkamp F, Godin K, Van Poppel H. Tolkach Y, et al. Korean J Urol. 2015 Feb;56(2):90-8. doi: 10.4111/kju.2015.56.2.90. Epub 2015 Jan 30. Korean J Urol. 2015. PMID: 25685295 Free PMC article. Review. - RAS/ERK pathway transcriptional regulation through ETS/AP-1 binding sites.
Hollenhorst PC. Hollenhorst PC. Small GTPases. 2012 Jul-Sep;3(3):154-8. doi: 10.4161/sgtp.19630. Epub 2012 Jun 1. Small GTPases. 2012. PMID: 22653334 Free PMC article. - Abiraterone in prostate cancer: a new angle to an old problem.
Stein MN, Goodin S, Dipaola RS. Stein MN, et al. Clin Cancer Res. 2012 Apr 1;18(7):1848-54. doi: 10.1158/1078-0432.CCR-11-1805. Epub 2012 Mar 26. Clin Cancer Res. 2012. PMID: 22451619 Free PMC article. Review.
References
- American Cancer Society: Cancer Facts & Figures 2007. 2007. www.acs.org.
- Lilja H, Ulmert D, Vickers AJ. Prostate-specific antigen and prostate cancer: prediction, detection and monitoring. Nat Rev Cancer. 2008;8:268–278. - PubMed
- Loeb S, Catalona WJ. Prostate-specific antigen in clinical practice. Cancer Lett. 2007;249:30–39. - PubMed
- Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases