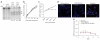Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases - PubMed (original) (raw)
doi: 10.1038/nbt1410. Epub 2008 Jun 29.
Jianbin Wang, Jeffrey C Miller, Yann Jouvenot, Kenneth A Kim, Olga Liu, Nathaniel Wang, Gary Lee, Victor V Bartsevich, Ya-Li Lee, Dmitry Y Guschin, Igor Rupniewski, Adam J Waite, Carmine Carpenito, Richard G Carroll, Jordan S Orange, Fyodor D Urnov, Edward J Rebar, Dale Ando, Philip D Gregory, James L Riley, Michael C Holmes, Carl H June
Affiliations
- PMID: 18587387
- PMCID: PMC3422503
- DOI: 10.1038/nbt1410
Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases
Elena E Perez et al. Nat Biotechnol. 2008 Jul.
Abstract
Homozygosity for the naturally occurring Delta32 deletion in the HIV co-receptor CCR5 confers resistance to HIV-1 infection. We generated an HIV-resistant genotype de novo using engineered zinc-finger nucleases (ZFNs) to disrupt endogenous CCR5. Transient expression of CCR5 ZFNs permanently and specifically disrupted approximately 50% of CCR5 alleles in a pool of primary human CD4(+) T cells. Genetic disruption of CCR5 provided robust, stable and heritable protection against HIV-1 infection in vitro and in vivo in a NOG model of HIV infection. HIV-1-infected mice engrafted with ZFN-modified CD4(+) T cells had lower viral loads and higher CD4(+) T-cell counts than mice engrafted with wild-type CD4(+) T cells, consistent with the potential to reconstitute immune function in individuals with HIV/AIDS by maintenance of an HIV-resistant CD4(+) T-cell population. Thus adoptive transfer of ex vivo expanded CCR5 ZFN-modified autologous CD4(+) T cells in HIV patients is an attractive approach for the treatment of HIV-1 infection.
Figures
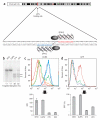
Figure 1
ZFN-mediated disruption of CCR5 and protection from HIV-1 infection in GHOST-CCR5 cells. (a) Schematic of the CCR5 coding region showing the genomic DNA sequences targeted by CCR5 ZFNs 215/224. (b) Level of targeted gene disruption in GHOST-CCR5 cells transduced with an Ad5/35 vector encoding ZFNs targeting either CCR5 or IL-2Rγ as assessed by the Surveyor assay (Supplementary Fig. 2). Lower migrating products (arrows) are a direct measure of ZFN-mediated gene disruption. NTD, nontransduced cells. (c) Decreased CCR5 surface expression measured by flow cytometry (light blue, IL2Rγ ZFN; green CCR5 ZFN-224; orange, CCR5 ZFN-215; dark blue, nontransduced cells; red, unstained cells). (d) Protection from HIV-1BAL measured by flow cytometry 48 h after HIV-1 challenge of CCR5 ZFN-215 and CCR5 ZFN-224-modified cells compared to IL2Rγ ZFN and control GHOST-CCR5 cells (red, IL2Rγ ZFN; green, CCR5 ZFN-224; blue, CCR5 ZFN-215). GFP fluorescence indicates HIV-1 entry and is plotted as average percent infected relative to positive control. MFI, mean fluorescence intensity. Histograms of CCR5 (c) and GFP expression (d) show one replicate for each condition; bar graphs below represent averages (±s.d.) of triplicates. Expression of CCR5 and HIV-1 infection frequency of CCR5 ZFN-treated cells is less than IL2Rγ ZFN or nontransduced cells (P < 0.001).
Figure 2
In vitro selection of CCR5-disrupted cells following HIV-1 challenge of the CD4+ T-cell line, PM1. (a) The level of ZFN-disrupted CCR5 alleles was determined at the indicated times post-HIV-1 challenge with R5-tropic HIV-1BAL or after mock HIV-1 infection. Disrupted CCR5 alleles remained at stable levels in mock-infected cultures but were enriched in the presence of HIV-1. Similar results were obtained in two experiments, each extending for over 2 months. (b) The genomic CCR5 ZFN target site in ZFN-treated PM1 cells at day 52 post-HIV-1 challenge was amplified, cloned and sequenced to confirm ZFN cleavage as the molecular basis of HIV-1 resistance. Sequence alignment revealed distinct ZFN-induced insertions and deletions within the target region of CCR5.
Figure 3
Enrichment of CCR5 ZFN-modified primary CD4+ T cells during in vitro HIV-1 challenge. (a) Primary CD4+ T cells from CCR5 wild-type anonymous healthy donor were transduced with Ad5/35 vector expressing CCR5 ZFN-215, ZFN-224 or GFP at MOI of 30 and 100; percentage of total alleles are indicated below each lane. (b) Population doubling rate for CCR5 ZFN- and GFP control-transduced CD4+ T cells (triangle, nontransduced; square, CCR5 ZFN-224; diamond, GFP transduced). (c) Enrichment of ZFN-215-transduced CD4+ T cells over time following in vitro challenge with CCR5-tropic HIV-1US1 compared to mock (square, HIV-1 infected; triangle, mock infected). An ~10% starting level of ZFN-disrupted CCR5 alleles was obtained by mixing Ad5/35-transduced CD4+ T cells from a 1 in 3 with unmodified CD4+ T cells. (d) Intranuclear 53BP1 immunostaining and epifluorescence microscopy 2 d after CD4+ T cells were transduced with Ad5/35 vectors expressing CCR5 ZFN pair 224, nontransduced (negative control) or 1 μM etoposide (positive control). Representative images are shown in the panels. (e) The mean (± s.d.) numbers of foci over time is shown following Ad5/35 vector transduction with CCR5 ZFN-224, GFP and nontransduced CD4+ T cells. Significant elevation in the number of foci was observed for CCR5 ZFN-224 treated cells on days 2 (P = 0.03) to 3 post treatment (P = 0.004, unpaired _t_-test, n = 4), whereas GFP-transduced cells were statistically indistinguishable from nontransduced control lymphocytes.
Figure 4
Reduction in viremia and selection for CCR5 ZFN-modified CD4+ T cells in the presence of HIV-1 challenge in vivo. (a) Experimental outline for adoptive transfer of modified CD4+ T cells and in vivo HIV-1 challenge in NOG mice. CCR5 ZFN-transduced CD4+ T-cell population pre-mix (day 3); CCR5 disruption level, 33%. Injected mixed samples baseline CCR5 disruption level 15% in control (mock infected) and 14% in HIV-infected group (day 7). (b) Level of ZFN-disrupted CCR5 alleles in CD4+ T cells isolated on day 40 from spleens of control or HIV-infected mice. Percent disruption indicated at base of each lane. One mouse from each group (HIV-infected mouse no. 8039 and control mouse no. 8022) excluded for later analysis due to inadequate CD4+ T-cell DNA recovery and purification. N.D., not determined. (c) Plot of in vivo disruption frequencies in spleens on day 40. Results for each group (n = 7) averaged and analyzed using an unpaired t-test with mean ± 95% confidence intervals indicated. (d–f) In an independent experiment, mice were engrafted with CCR5 ZFN-transduced CD4+ T cells (51% disruption) or GFP-transduced cells (mock) and followed for 50 d post HIV-1 infection. Enrichment for CCR5-disrupted CD4+ T cells in peripheral blood on day 50 post infection (Surveyor assay); lower migrating products (arrows) are a direct measure of ZFN-mediated gene disruption (d). Plasma viremia in mice day 10 post infection. HIV-1 viral RNA (copies/ml) is plotted for the individual mice; the mean ± 95% confidence interval is shown (e). The CCR5 ZFN-treated mice had a significantly lower viral load (P < 0.001; Mann Whitney test). Engraftment of CD4+ T cells in peripheral blood from days 10 to 50 post infection. The CD4+ T-cell counts (Trucount assay) for the mice engrafted with CCR5 ZFN-(solid symbols) and GFP-modified cells (open symbols) are plotted (f). Mice engrafted with CCR5 ZFN-treated cells had higher CD4+ T-cell counts on days 30–50 post infection (P = 0.04).
Similar articles
- Engineering HIV-resistant human CD4+ T cells with CXCR4-specific zinc-finger nucleases.
Wilen CB, Wang J, Tilton JC, Miller JC, Kim KA, Rebar EJ, Sherrill-Mix SA, Patro SC, Secreto AJ, Jordan AP, Lee G, Kahn J, Aye PP, Bunnell BA, Lackner AA, Hoxie JA, Danet-Desnoyers GA, Bushman FD, Riley JL, Gregory PD, June CH, Holmes MC, Doms RW. Wilen CB, et al. PLoS Pathog. 2011 Apr;7(4):e1002020. doi: 10.1371/journal.ppat.1002020. Epub 2011 Apr 14. PLoS Pathog. 2011. PMID: 21533216 Free PMC article. - Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo.
Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, Crooks GM, Kohn DB, Gregory PD, Holmes MC, Cannon PM. Holt N, et al. Nat Biotechnol. 2010 Aug;28(8):839-47. doi: 10.1038/nbt.1663. Epub 2010 Jul 2. Nat Biotechnol. 2010. PMID: 20601939 Free PMC article. - Efficient clinical scale gene modification via zinc finger nuclease-targeted disruption of the HIV co-receptor CCR5.
Maier DA, Brennan AL, Jiang S, Binder-Scholl GK, Lee G, Plesa G, Zheng Z, Cotte J, Carpenito C, Wood T, Spratt SK, Ando D, Gregory P, Holmes MC, Perez EE, Riley JL, Carroll RG, June CH, Levine BL. Maier DA, et al. Hum Gene Ther. 2013 Mar;24(3):245-58. doi: 10.1089/hum.2012.172. Epub 2013 Mar 6. Hum Gene Ther. 2013. PMID: 23360514 Free PMC article. - Chemokine receptor 5 knockout strategies.
Cannon P, June C. Cannon P, et al. Curr Opin HIV AIDS. 2011 Jan;6(1):74-9. doi: 10.1097/COH.0b013e32834122d7. Curr Opin HIV AIDS. 2011. PMID: 21242897 Free PMC article. Review. - Pre-clinical modeling of CCR5 knockout in human hematopoietic stem cells by zinc finger nucleases using humanized mice.
Hofer U, Henley JE, Exline CM, Mulhern O, Lopez E, Cannon PM. Hofer U, et al. J Infect Dis. 2013 Nov;208 Suppl 2(Suppl 2):S160-4. doi: 10.1093/infdis/jit382. J Infect Dis. 2013. PMID: 24151324 Free PMC article. Review.
Cited by
- Nonhomologous end joining-mediated gene replacement in plant cells.
Weinthal DM, Taylor RA, Tzfira T. Weinthal DM, et al. Plant Physiol. 2013 May;162(1):390-400. doi: 10.1104/pp.112.212910. Epub 2013 Mar 18. Plant Physiol. 2013. PMID: 23509176 Free PMC article. - CRISPR-Cas9 Technology for the Creation of Biological Avatars Capable of Modeling and Treating Pathologies: From Discovery to the Latest Improvements.
Nasrallah A, Sulpice E, Kobaisi F, Gidrol X, Rachidi W. Nasrallah A, et al. Cells. 2022 Nov 15;11(22):3615. doi: 10.3390/cells11223615. Cells. 2022. PMID: 36429042 Free PMC article. Review. - Cas-analyzer: an online tool for assessing genome editing results using NGS data.
Park J, Lim K, Kim JS, Bae S. Park J, et al. Bioinformatics. 2017 Jan 15;33(2):286-288. doi: 10.1093/bioinformatics/btw561. Epub 2016 Aug 24. Bioinformatics. 2017. PMID: 27559154 Free PMC article. - Humanized Mice for the Evaluation of Novel HIV-1 Therapies.
Abeynaike S, Paust S. Abeynaike S, et al. Front Immunol. 2021 Apr 1;12:636775. doi: 10.3389/fimmu.2021.636775. eCollection 2021. Front Immunol. 2021. PMID: 33868262 Free PMC article. Review.
References
- Deng HK, et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. - PubMed
- Alkhatib G, et al. Cc Ckrs: A Rantes, Mip-1 Alpha, Mip-1 Beta Receptor As A Fusion Cofactor for Macrophage-Tropic HIV-1. Science. 1996;272:1955–1958. - PubMed
- Liu R, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. - PubMed
- Samson M, et al. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. - PubMed
- Huang YX, et al. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat. Med. 1996;2:1240–1243. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K08 AI062468/AI/NIAID NIH HHS/United States
- T32 HL007775/HL/NHLBI NIH HHS/United States
- U19 AI082628/AI/NIAID NIH HHS/United States
- K08AI062468/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials