Cyclin-specific control of ribosomal DNA segregation - PubMed (original) (raw)
Cyclin-specific control of ribosomal DNA segregation
Matt Sullivan et al. Mol Cell Biol. 2008 Sep.
Abstract
Following chromosome duplication in S phase of the cell cycle, the sister chromatids are linked by cohesin. At the onset of anaphase, separase cleaves cohesin and thereby initiates sister chromatid separation. Separase activation results from the destruction of its inhibitor, securin, which is triggered by a ubiquitin ligase called the anaphase-promoting complex (APC). Here, we show in budding yeast that securin destruction and, thus, separase activation are not sufficient for the efficient segregation of the repetitive ribosomal DNA (rDNA). We find that rDNA segregation also requires the APC-mediated destruction of the S-phase cyclin Clb5, an activator of the protein kinase Cdk1. Mutations that prevent Clb5 destruction are lethal and cause defects in rDNA segregation and DNA synthesis. These defects are distinct from the mitotic-exit defects caused by stabilization of the mitotic cyclin Clb2, emphasizing the importance of cyclin specificity in the regulation of late-mitotic events. Efficient rDNA segregation, both in mitosis and meiosis, also requires APC-dependent destruction of Dbf4, an activator of the protein kinase Cdc7. We speculate that the dephosphorylation of Clb5-specific Cdk1 substrates and Dbf4-Cdc7 substrates drives the resolution of rDNA in early anaphase. The coincident destruction of securin, Clb5, and Dbf4 coordinates bulk chromosome segregation with segregation of rDNA.
Figures
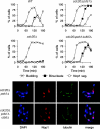
FIG. 1.
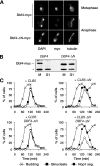
APCCdc20-dependent degradation of securin and Clb5 allows rDNA segregation. Wild-type (WT), _cdc20_Δ, _cdc20_Δ _pds1_Δ, and _cdc20_Δ _pds1_Δ _clb5_Δ yeast expressing GAL-CDC20 were arrested in G1 with α-factor and then released into fresh dextrose media to repress CDC20 transcription. Samples were collected every 20 min for analysis of budding, binucleate formation (bulk chromosome segregation, assessed by DAPI [4′,6-diamidino-2-phenylindole] staining), and Nop1 segregation (a marker of nucleolar rDNA segregation) (top panel). The bottom panel shows representative images of _cdc20_Δ _pds1_Δ and _cdc20_Δ _pds1_Δ _clb5_Δ yeast 140 min after release from G1.
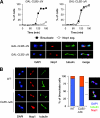
FIG. 2.
Clb5 inhibits rDNA segregation. (A) Cells carrying either GAL-CLB5_-Δ_N or GAL-CLB2_-Δ_N were arrested in G1 with α-factor and then released into fresh galactose media to induce cyclin expression. Samples were collected every 15 min for analysis of binucleate formation and Nop1 segregation. Representative images are shown of GAL-CLB5_-Δ_N yeast and GAL-CLB2_-Δ_N yeast 120 min after release from G1. (B) Asynchronous CLB5 (WT) and CLB5_-Δ_db cells were collected and binucleates analyzed for the number of segregated and unsegregated Nop1 masses. Representative images are shown (left).
FIG. 3.
CLB5-ΔN is lethal and causes an rDNA segregation delay. (A) Wild-type or _rDNA_Δ cells, carrying GAL-SIC1 and an extra copy of the CLB5 or CLB5_-Δ_N gene (expressed from the CLB5 promoter), were plated on either galactose or dextrose media. (B) GAL-SIC1 cells carrying an extra copy of CLB5 or CLB5_-Δ_N were arrested in G1 by α-factor treatment and released into fresh dextrose media to repress SIC1 transcription. Samples were taken every 20 min, and the levels of Clb5 and Clb2 were analyzed by Western blotting. (C) Procedure was as in panel B, but α-factor was added 90 min after release to arrest cells in the following G1. Samples were collected every 15 min for analysis of budding, binucleate formation, and Nop1 segregation. Images on the right show representative cells 140 min after release from G1.
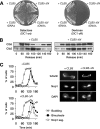
FIG. 4.
CLB5_-Δ_N causes DNA replication defects. (A) CLB5 and CLB5_-Δ_N cells were released from G1 arrest in the absence of SIC1 as in Fig. 3B, and flow cytometry was used to analyze cellular DNA content. (B) Procedure was as in panel A but with MCM4-GFP cells, which were analyzed for budding, binucleates, and the localization of the Mcm4-GFP protein.
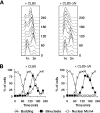
FIG. 5.
Stabilization of Dbf4 contributes to the rDNA segregation delay. (A) Representative images of Dbf4-myc in metaphase or anaphase from wild-type or DBF4_-Δ_N cells. (B) DBF4 and DBF4_-Δ_N cells were arrested in M phase by nocodazole treatment or G1 by α-factor treatment, and the levels of Dbf4 were analyzed. The asterisk indicates a nonspecific background band recognized by the anti-myc antibody. (C) DBF4 and DBF4-ΔN cells, carrying an extra copy of either CLB5 or CLB5_-Δ_N, were released from a G1 arrest as in Fig. 3C, and α-factor was re-added 80 min after release to arrest cells in the following G1. Samples were collected every 20 min for analysis of budding, binucleate formation, and Nop1 segregation.
FIG. 6.
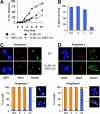
Meiotic rDNA segregation fails in the presence of stabilized Dbf4 and Clb5. Diploid DBF4 and DBF4_-Δ_N SK1 cells, with or without pDMC1-CLB5_-Δ_N, were followed through synchronous meiosis. Chromosome divisions were scored (A), as were the survival rates of resulting spores (B) and the segregation patterns of Nop1 through both anaphase I (C) and anaphase II (D). Histograms are labeled WT (wild type), d (DBF4_-Δ_N), c (pDMC1-CLB5_-Δ_N), and dc (DBF4-ΔN with pDMC1-CLB5_-Δ_N).
FIG. 7.
Model of regulatory mechanisms governing chromosome segregation in budding yeast. Bulk chromosome separation occurs when cohesin is cleaved by separase, the activation of which results from the APCCdc20-dependent destruction of securin. Efficient removal of rDNA cohesion also requires the dephosphorylation of Clb5-specific Cdk1 targets, which results from the activation of Cdc14 by separase and the destruction of Clb5 via APCCdc20. By uncertain mechanisms, APCCdc20-dependent Dbf4 destruction also contributes. Destruction of Clb2 allows dephosphorylation of its targets after anaphase, leading to spindle disassembly and mitotic exit.
References
- Arias, E. E., and J. C. Walter. 2007. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 21497-518. - PubMed
- Bloom, J., and F. R. Cross. 2007. Multiple levels of cyclin specificity in cell-cycle control. Nat. Rev. Mol. Cell Biol. 8149-160. - PubMed
- Chen, C. T., M. P. Peli-Gulli, V. Simanis, and D. McCollum. 2006. S. pombe FEAR protein orthologs are not required for release of Clp1/Flp1 phosphatase from the nucleolus during mitosis. J. Cell Sci. 1194462-4466. - PubMed
- Cross, F. R., M. Yuste-Rojas, S. Gray, and M. D. Jacobson. 1999. Specialization and targeting of B-type cyclins. Mol. Cell 411-19. - PubMed
- D'Amours, D., F. Stegmeier, and A. Amon. 2004. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell 117455-469. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous