TDRD3, a novel Tudor domain-containing protein, localizes to cytoplasmic stress granules - PubMed (original) (raw)
. 2008 Oct 1;17(19):3055-74.
doi: 10.1093/hmg/ddn203. Epub 2008 Jul 15.
Affiliations
- PMID: 18632687
- PMCID: PMC2536506
- DOI: 10.1093/hmg/ddn203
TDRD3, a novel Tudor domain-containing protein, localizes to cytoplasmic stress granules
Isabelle Goulet et al. Hum Mol Genet. 2008.
Abstract
Our previous work has demonstrated that the Tudor domain of the 'survival of motor neuron' protein and the Tudor domain-containing protein 3 (TDRD3) are highly similar and that they both have the ability to interact with arginine-methylated polypeptides. TDRD3 has been identified among genes whose overexpression has a strong predictive value for poor prognosis of estrogen receptor-negative breast cancers, although its precise function remains unknown. TDRD3 is a modular protein, and in addition to its Tudor domain, it harbors a putative nucleic acid recognition motif and a ubiquitin-associated domain. We report here that TDRD3 localizes predominantly to the cytoplasm, where it co-sediments with the fragile X mental retardation protein on actively translating polyribosomes. We also demonstrate that TDRD3 accumulates into stress granules (SGs) in response to various cellular stresses. Strikingly, the Tudor domain of TDRD3 was found to be both required and sufficient for its recruitment to SGs, and the methyl-binding surface in the Tudor domain is important for this process. Pull down experiments identified five novel TDRD3 interacting partners, most of which are potentially methylated RNA-binding proteins. Our findings revealed that two of these proteins, SERPINE1 mRNA-binding protein 1 and DEAD/H box-3 (a gene often deleted in Sertoli-cell-only syndrome), are also novel constituents of cytoplasmic SGs. Taken together, we report the first characterization of TDRD3 and its functional interaction with at least two proteins implicated in human genetic diseases and present evidence supporting a role for arginine methylation in the regulation of SG dynamics.
Figures
Figure 1.
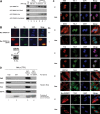
Genomic organization of the human TDRD3 locus and schematic representation of the TDRD3 protein. Analysis of the human TDRD3 gene locus revealed 14 exons spanning over ∼176.5 kb of genomic DNA, encoding a 2.9-kb mRNA transcript (A). The sequence of exons is displayed with capital letters above the amino acid sequences they encode. Only short sequences around the intron boundaries are shown with small letters. All of the exon-intron junctions (shown in black boxes) conform to the GU-AG general consensus. The start and stop codons are in bold, whereas the Kozak sequence and the polyadenylation signal are underlined. Both nucleotides and amino acids are numbered starting from the ATG codon (B). The encoded TDRD3 protein consists of 744 amino acids arranged in a modular fashion. Data obtained from the NCBI Conserved Domains and the Protein Family (Pfam version 22.0) databases predict the presence of three structural domains in TDRD3: a DUF1767/OB-fold (amino acids 13–169), a UBA (amino acids 286–328) and a Tudor (amino acids 651–710) domain (see text for details) (C).
Figure 2.
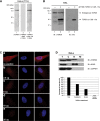
A polyclonal antibody against the novel TDRD3 protein. A HeLa total cell extract was immnublotted using our affinity-purified anti-TDRD3 polyclonal antibody (lane 1). Alternatively, immunoblots were performed using the TDRD3 antibody preincubated with a 200-fold molar excess of either the antigenic peptide (lane 2) or an unrelated peptide (ctrl; lane 3) (A). In vitro transcription and translation reactions were programmed to express a hexahistidine-tagged truncated version of TDRD3 (ΔN328). Control (−) and programmed (+) reactions were resolved by SDS–PAGE, transferred to a PVDF membrane, and immunoblotted with either TDRD3 (lanes 1 and 2) or His (lanes 3 and 4) antibodies (B). Actively growing HeLa cells were labeled for immunofluorescence with our TDRD3 antibody (a). Alternatively, TDRD3 antibodies were preincubated with an increasing amount (2.5–25 g) of the immunogenic peptide for 1 h on ice prior to immunofluorescence staining (b–e) (C). HeLa cells were fractionated using the Qproteome Nuclear Protein Kit (Qiagen). 2.5% of each fraction was resolved by SDS–PAGE, transferred to PVDF and immunoblotted with affinity-purified anti-TDRD3. Cellular fractionation efficiency was assessed using antibodies against the nuclear protein EWS and the cytoplasmic protein GAPDH. Total (T), cytoplasmic (C), nuclear (N) and insoluble nuclear protein (NI) fractions are shown (top). Quantification of the relative abundance of TDRD3 in each fraction is presented in a bar graph (bottom) (D).
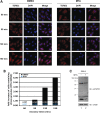
Figure 3.
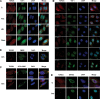
TDRD3 localizes to cytoplasmic stress granules during stress response. HeLa cells cultured on glass cover slips were left untreated (C) or exposed to oxidative stress (Ars; 0.5 m
m
sodium arsenite for 30 min), heat shock (HS; 43°C for 30 min) or high-osmolarity medium (Osm; 1 M sorbitol in DMEM for 1 h followed by a 30-min recovery in normal DMEM). The cells were fixed and immunostained with TDRD3 and TIA-1 antibodies to detect the endogenous proteins (A). Cells were stressed with 0.5 m
m
sodium arsenite (Ars) for 30 min, followed by immunostaining of both TDRD3 and FMRP endogenous proteins (B). To visualize G3BP, a GFP fusion construct was transfected into cells 24 h prior to immunofluorescence analysis. Cells were left untreated (C) or exposed to oxidative stress (Ars) for 30 min, followed by immunostaining for endogenous TDRD3 (C). Cells were stressed with 0.5 m
m
sodium arsenite for the indicated time length before their fixation and double immunofluorescence with TDRD3 and TIA-1 antibodies (D). HeLa cells were either left untreated (C) or stressed for 30 min with 0.5 m
m
sodium arsenite (Ars). After fixation, endogenous TDRD3 and GW182 (a marker of P-bodies) were detected by fluorescence microscopy (E).
Figure 4.
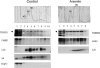
TDRD3 is associated with polyribosomes in HeLa cells. Cytoplasmic extracts were prepared from HeLa cells grown in normal conditions and centrifuged on a 10–60% w/w linear sucrose gradient. Fractions were collected and analyzed by western blot with antibodies against the ribosomal S6 protein and FMRP (as positive controls), TDRD3, and Hsp72 (as a negative control) (Control panel). Cytoplasmic extracts from HeLa cells treated with 0.5 m
m
sodium arsenite for 30 min were analyzed in parallel and the collected fractions were analyzed by western blot using the ribosomal L28 protein as positive control (Arsenite panel). Fractions from the top to the bottom of the gradient are shown from left to right. The positions of free small (40S) and large (60S) ribosomal subunits, monosomes (80S), and polysomes are indicated in each profile. The band corresponding to TDRD3 is indicated by a ‘dot’ on the side of the respective panels. Additional bands detected on the immunoblots represent non-specific reactivity with our polyclonal antibody.
Figure 5.
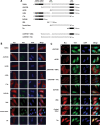
The Tudor domain of TDRD3 is both required and sufficient for its recruitment to stress granules. Diagram showing the various myc epitope-tagged TDRD3 deletion mutants used in this study (A). HeLa cells were transiently transfected with each deletion mutant. Twenty-four hours post-transfection, cells were left untreated and labeled for immunofluorescence with TIA-1 and myc antibodies to detect endogenous TIA-1 protein and recombinant myc-tagged proteins (B). Alternatively, transfected cells were treated with 0.5 m
m
sodium arsenite for 30 min. Indirect immunofluorescence staining was performed as described above (C). HeLa cells were transiently transfected with constructs expressing recombinant myc-tagged Tudor domain of SMN, SPF30 or TDRD3. Twenty-four hours post-transfection, cells were treated with 0.5 m
m
sodium arsenite for 30 min. Indirect immunofluorescence staining was performed using myc and TIA-1 antibodies (D). Total cell lysates of HeLa cells transiently expressing myc-tagged TDRD3 deletion mutants, as well as SMN and SPF30 Tudor domains, were immunoblotted with myc antibodies to confirm expression (E).
Figure 5.
The Tudor domain of TDRD3 is both required and sufficient for its recruitment to stress granules. Diagram showing the various myc epitope-tagged TDRD3 deletion mutants used in this study (A). HeLa cells were transiently transfected with each deletion mutant. Twenty-four hours post-transfection, cells were left untreated and labeled for immunofluorescence with TIA-1 and myc antibodies to detect endogenous TIA-1 protein and recombinant myc-tagged proteins (B). Alternatively, transfected cells were treated with 0.5 m
m
sodium arsenite for 30 min. Indirect immunofluorescence staining was performed as described above (C). HeLa cells were transiently transfected with constructs expressing recombinant myc-tagged Tudor domain of SMN, SPF30 or TDRD3. Twenty-four hours post-transfection, cells were treated with 0.5 m
m
sodium arsenite for 30 min. Indirect immunofluorescence staining was performed using myc and TIA-1 antibodies (D). Total cell lysates of HeLa cells transiently expressing myc-tagged TDRD3 deletion mutants, as well as SMN and SPF30 Tudor domains, were immunoblotted with myc antibodies to confirm expression (E).
Figure 6.
Direct contact with methylated arginines contributes to TDRD3 relocalization to SGs. Biotinylated (RG4) peptides containing either unmethylated arginines or symmetrically dimethylated arginines (sDMA) were bound to streptavidin-agarose and used as affinity columns to measure the binding of purified GST fusion SMN and SMN E134K (as controls) and/or TDRD3 and TDRD3 E691K Tudor domains. The bound GST-Tdr fusion proteins were resolved by SDS-PAGE, transferred to a PVDF membrane, and detected by immunoblotting using GST antibodies (A). HeLa cells were transiently transfected with constructs expressing recombinant myc-tagged wild type or mutated (E691K) TDRD3 Tudor domain. Twenty-four hours post-transfection, cells were treated with 0.5 m
m
sodium arsenite for 30 min. Indirect immunofluorescence staining was performed using myc and TIA-1 antibodies (B). HeLa cells were either left untreated (C) or treated with 0.5 m
m
sodium arsenite for 30 min (Ars). Cells were then labeled for immunofluorescence with EWS (a and b), DDX3 (c and d), FUS (e and f), or EEF1A1 (g and h) in combination with TIA-1 antibodies to detect endogenous proteins. HeLa cells transiently transfected to express myc-tagged SERBP1 (i and j) were either left untreated (C) or stressed as described above (Ars), before being immunostained with TDRD3 and myc (SERBP1) antibodies (C). HeLa cells from 2× 150 mm plates were lysed and incubated with purified recombinant GST (as a control), GST-TDRD3-Tdr or GST-TDRD3-Tdr E691K proteins coupled to glutathione-agarose. The retained proteins were resolved by SDS-PAGE, transferred to PVDF, and immunoblotted with the specified antibodies to confirm mass spectrometry identifications. The membrane was stained with Ponceau Red prior to immunoblotting, in order to show the GST-fusion proteins (D).
Figure 7.
Arginine methylation and SG dynamics. HeLa cells grown on cover slips were stressed with 0.5 m
m
sodium arsenite for 30 min. Cells were then returned to normal growth media and prepared for TDRD3/TIA-1 immunostaining after the indicated incubation periods (A). The experiment was repeated twice using different batches of cells. At least 500 cells were counted for each time point and condition. Fold increase of SG-positive cells in the MTA-treated cells when compared the DMSO-treated cells is depicted in a bar graph (B). Cell extracts prepared from HeLa cells grown in the presence (+) or the absence (−) of the general methylation inhibitor MTA were immunoblotted with the SYM10 antibody to confirm the reduction in steady-state arginine methylation levels. The same extracts were immunoblotted with actin antibodies to control for equal loading (C).
Similar articles
- Tdrd3 is a novel stress granule-associated protein interacting with the Fragile-X syndrome protein FMRP.
Linder B, Plöttner O, Kroiss M, Hartmann E, Laggerbauer B, Meister G, Keidel E, Fischer U. Linder B, et al. Hum Mol Genet. 2008 Oct 15;17(20):3236-46. doi: 10.1093/hmg/ddn219. Epub 2008 Jul 28. Hum Mol Genet. 2008. PMID: 18664458 - Structural plasticity of the TDRD3 Tudor domain probed by a fragment screening hit.
Liu J, Zhang S, Liu M, Liu Y, Nshogoza G, Gao J, Ma R, Yang Y, Wu J, Zhang J, Li F, Ruan K. Liu J, et al. FEBS J. 2018 Jun;285(11):2091-2103. doi: 10.1111/febs.14469. Epub 2018 Apr 24. FEBS J. 2018. PMID: 29645362 - Arginine methylation of USP9X promotes its interaction with TDRD3 and its anti-apoptotic activities in breast cancer cells.
Narayanan N, Wang Z, Li L, Yang Y. Narayanan N, et al. Cell Discov. 2017 Jan 3;3:16048. doi: 10.1038/celldisc.2016.48. eCollection 2017. Cell Discov. 2017. PMID: 28101374 Free PMC article. - Type IA topoisomerases can be "magicians" for both DNA and RNA in all domains of life.
Ahmad M, Xu D, Wang W. Ahmad M, et al. RNA Biol. 2017 Jul 3;14(7):854-864. doi: 10.1080/15476286.2017.1330741. Epub 2017 May 23. RNA Biol. 2017. PMID: 28534707 Free PMC article. Review. - Tudor domain-containing proteins of Drosophila melanogaster.
Ying M, Chen D. Ying M, et al. Dev Growth Differ. 2012 Jan;54(1):32-43. doi: 10.1111/j.1440-169x.2011.01308.x. Dev Growth Differ. 2012. PMID: 23741747 Review.
Cited by
- mRNP granules. Assembly, function, and connections with disease.
Buchan JR. Buchan JR. RNA Biol. 2014;11(8):1019-30. doi: 10.4161/15476286.2014.972208. RNA Biol. 2014. PMID: 25531407 Free PMC article. Review. - Who's In and Who's Out-Compositional Control of Biomolecular Condensates.
Ditlev JA, Case LB, Rosen MK. Ditlev JA, et al. J Mol Biol. 2018 Nov 2;430(23):4666-4684. doi: 10.1016/j.jmb.2018.08.003. Epub 2018 Aug 9. J Mol Biol. 2018. PMID: 30099028 Free PMC article. Review. - The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex.
Hilliker A, Gao Z, Jankowsky E, Parker R. Hilliker A, et al. Mol Cell. 2011 Sep 16;43(6):962-72. doi: 10.1016/j.molcel.2011.08.008. Mol Cell. 2011. PMID: 21925384 Free PMC article. - DDX3 Interacts with Influenza A Virus NS1 and NP Proteins and Exerts Antiviral Function through Regulation of Stress Granule Formation.
Thulasi Raman SN, Liu G, Pyo HM, Cui YC, Xu F, Ayalew LE, Tikoo SK, Zhou Y. Thulasi Raman SN, et al. J Virol. 2016 Jan 20;90(7):3661-75. doi: 10.1128/JVI.03010-15. J Virol. 2016. PMID: 26792746 Free PMC article. - Reading Cancer: Chromatin Readers as Druggable Targets for Cancer Treatment.
Mio C, Bulotta S, Russo D, Damante G. Mio C, et al. Cancers (Basel). 2019 Jan 9;11(1):61. doi: 10.3390/cancers11010061. Cancers (Basel). 2019. PMID: 30634442 Free PMC article. Review.
References
- Pawson T. Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell. 2004;116:191–203. - PubMed
- Lee D.Y., Teyssier C., Strahl B.D., Stallcup M.R. Role of protein methylation in regulation of transcription. Endocr. Rev. 2005;26:147–170. - PubMed
- Bedford M.T., Richard S. Arginine methylation an emerging regulator of protein function. Mol. Cell. 2005;18:263–272. - PubMed
- Boisvert F.M., Chenard C.A., Richard S. Protein interfaces in signaling regulated by arginine methylation. Sci. STKE. 2005;2005:re2. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous