Blocking primers to enhance PCR amplification of rare sequences in mixed samples - a case study on prey DNA in Antarctic krill stomachs - PubMed (original) (raw)
Blocking primers to enhance PCR amplification of rare sequences in mixed samples - a case study on prey DNA in Antarctic krill stomachs
Hege Vestheim et al. Front Zool. 2008.
Abstract
Background: Identification of DNA sequence diversity is a powerful means for assessing the species present in environmental samples. The most common molecular strategies for estimating taxonomic composition depend upon PCR with universal primers that amplify an orthologous DNA region from a range of species. The diversity of sequences within a sample that can be detected by universal primers is often compromised by high concentrations of some DNA templates. If the DNA within the sample contains a small number of sequences in relatively high concentrations, then less concentrated sequences are often not amplified because the PCR favours the dominant DNA types. This is a particular problem in molecular diet studies, where predator DNA is often present in great excess of food-derived DNA.
Results: We have developed a strategy where a universal PCR simultaneously amplifies DNA from food items present in DNA purified from stomach samples, while the predator's own DNA is blocked from amplification by the addition of a modified predator-specific blocking primer. Three different types of modified primers were tested out; one annealing inhibiting primer overlapping with the 3' end of one of the universal primers, another annealing inhibiting primer also having an internal modification of five dI molecules making it a dual priming oligo, and a third elongation arrest primer located between the two universal primers. All blocking primers were modified with a C3 spacer. In artificial PCR mixtures, annealing inhibiting primers proved to be the most efficient ones and this method reduced predator amplicons to undetectable levels even when predator template was present in 1000 fold excess of the prey template. The prey template then showed strong PCR amplification where none was detectable without the addition of blocking primer. Our method was applied to identifying the winter food of one of the most abundant animals in the world, the Antarctic krill, Euphausia superba. Dietary item DNA was PCR amplified from a range of species in krill stomachs for which we had no prior sequence knowledge.
Conclusion: We present a simple, robust and cheap method that is easily adaptable to many situations where a rare DNA template is to be PCR amplified in the presence of a higher concentration template with identical PCR primer binding sites.
Figures
Figure 1
Screening methods. Schematic illustration of different ways to screen away a dominating DNA template in a PCR mixture. 1. Universal primers amplifying target and non-target DNA and removal of non-target DNA prior or after PCR with restriction enzymes. 2. Species-specific PCR primers amplifying only target-DNA. 3. Group-specific PCR primers amplifying groups excluding non-target sequence. 4. Blocking non-target amplification, only target sequences amplified using universal PCR primers a) annealing inhibiting blocking primer, b) elongation arrest blocking primer c) annealing inhibiting DPO blocking primer.
Figure 2
Primers. A 203 bp region of Euphausia superba 28S sequence showing the location of the different primers applied in this study.
Figure 3
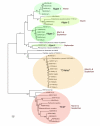
Sequence similarity tree. Sequence similarity tree of sequences amplified from krill stomachs and related sequences. The krill stomach sequences are named so as the first three letters means sampling date, i.e. M24 = March 24. 2007, S17 and S20 = September 17. and September 20. 2007, St. means Stomach number and the number in parenthesis equals the number of identical sequences of each different sequence found in the different stomachs. The tree is unrooted.
Similar articles
- Universal and blocking primer mismatches limit the use of high-throughput DNA sequencing for the quantitative metabarcoding of arthropods.
Piñol J, Mir G, Gomez-Polo P, Agustí N. Piñol J, et al. Mol Ecol Resour. 2015 Jul;15(4):819-30. doi: 10.1111/1755-0998.12355. Epub 2014 Dec 23. Mol Ecol Resour. 2015. PMID: 25454249 - Group-specific polymerase chain reaction for DNA-based analysis of species diversity and identity in dietary samples.
Jarman SN, Deagle BE, Gales NJ. Jarman SN, et al. Mol Ecol. 2004 May;13(5):1313-22. doi: 10.1111/j.1365-294X.2004.02109.x. Mol Ecol. 2004. PMID: 15078466 - [Polymerase chain reaction, cold probes and clinical diagnosis].
Haras D, Amoros JP. Haras D, et al. Sante. 1994 Jan-Feb;4(1):43-52. Sante. 1994. PMID: 7909267 Review. French. - [Quantitative PCR in the diagnosis of Leishmania].
Mortarino M, Franceschi A, Mancianti F, Bazzocchi C, Genchi C, Bandi C. Mortarino M, et al. Parassitologia. 2004 Jun;46(1-2):163-7. Parassitologia. 2004. PMID: 15305709 Review. Italian.
Cited by
- Comprehensive coverage of human last meal components revealed by a forensic DNA metabarcoding approach.
Schneider J, Mas-Carrió E, Jan C, Miquel C, Taberlet P, Michaud K, Fumagalli L. Schneider J, et al. Sci Rep. 2021 Apr 23;11(1):8876. doi: 10.1038/s41598-021-88418-x. Sci Rep. 2021. PMID: 33893381 Free PMC article. - Preliminary analysis of New Zealand scampi (Metanephrops challengeri) diet using metabarcoding.
van der Reis AL, Laroche O, Jeffs AG, Lavery SD. van der Reis AL, et al. PeerJ. 2018 Sep 20;6:e5641. doi: 10.7717/peerj.5641. eCollection 2018. PeerJ. 2018. PMID: 30258728 Free PMC article. - Variation in diet composition and its relation to gut microbiota in a passerine bird.
Schmiedová L, Tomášek O, Pinkasová H, Albrecht T, Kreisinger J. Schmiedová L, et al. Sci Rep. 2022 Mar 8;12(1):3787. doi: 10.1038/s41598-022-07672-9. Sci Rep. 2022. PMID: 35260644 Free PMC article. - DNA metabarcoding reveals a broad dietary range for Tasmanian devils introduced to a naive ecosystem.
McLennan EA, Wise P, Lee AV, Grueber CE, Belov K, Hogg CJ. McLennan EA, et al. Ecol Evol. 2022 May 19;12(5):e8936. doi: 10.1002/ece3.8936. eCollection 2022 May. Ecol Evol. 2022. PMID: 35600680 Free PMC article. - The treasure inside barley seeds: microbial diversity and plant beneficial bacteria.
Bziuk N, Maccario L, Straube B, Wehner G, Sørensen SJ, Schikora A, Smalla K. Bziuk N, et al. Environ Microbiome. 2021 Oct 28;16(1):20. doi: 10.1186/s40793-021-00389-8. Environ Microbiome. 2021. PMID: 34711269 Free PMC article.
References
- Loeb V, Siegel V, HolmHansen O, Hewitt R, Fraser W, Trivelpiece W, Trivelpiece S. Effects of sea-ice extent and krill or salp dominance on the Antarctic food web. Nature. 1997;387:897–900. doi: 10.1038/43174. - DOI
- Heywood BG, Brierley AS, Gull SF. A quantified Bayesian maximum entropy estimate of Antarctic krill abundance across the Scotia Sea and in small-scale management units from the CCAMLR-2000 survey. CCAMLR Science. 2006;13:97–116.
- Croxall JP, Nicol S. Management of Southern Ocean fisheries: global forces and future sustainability. Antarctic Science. 2004;16:569–584. doi: 10.1017/S0954102004002330. - DOI
- Marr JSW. Discovery report. 1962.
- Schmidt K, Atkinson A, Petzke KJ, Voss M, Pond DW. Protozoans as a food source for Antarctic krill, Euphausia superba: Complementary insights from stomach content, fatty acids, and stable isotopes. Limnology and Oceanography. 2006;51:2409–2427.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous