Structural dissection of a gating mechanism preventing misactivation of ubiquitin by NEDD8's E1 - PubMed (original) (raw)
. 2008 Aug 26;47(34):8961-9.
doi: 10.1021/bi800604c. Epub 2008 Jul 25.
Affiliations
- PMID: 18652489
- PMCID: PMC2587436
- DOI: 10.1021/bi800604c
Structural dissection of a gating mechanism preventing misactivation of ubiquitin by NEDD8's E1
Judith Souphron et al. Biochemistry. 2008.
Abstract
Post-translational covalent modification by ubiquitin and ubiquitin-like proteins (UBLs) is a major eukaryotic mechanism for regulating protein function. In general, each UBL has its own E1 that serves as the entry point for a cascade. The E1 first binds the UBL and catalyzes adenylation of the UBL's C-terminus, prior to promoting UBL transfer to a downstream E2. Ubiquitin's Arg 72, which corresponds to Ala72 in the UBL NEDD8, is a key E1 selectivity determinant: swapping ubiquitin and NEDD8 residue 72 identity was shown previously to swap their E1 specificity. Correspondingly, Arg190 in the UBA3 subunit of NEDD8's heterodimeric E1 (the APPBP1-UBA3 complex), which corresponds to a Gln in ubiquitin's E1 UBA1, is a key UBL selectivity determinant. Here, we dissect this specificity with biochemical and X-ray crystallographic analysis of APPBP1-UBA3-NEDD8 complexes in which NEDD8's residue 72 and UBA3's residue 190 are substituted with different combinations of Ala, Arg, or Gln. APPBP1-UBA3's preference for NEDD8's Ala72 appears to be indirect, due to proper positioning of UBA3's Arg190. By contrast, our data are consistent with direct positive interactions between ubiquitin's Arg72 and an E1's Gln. However, APPBP1-UBA3's failure to interact with a UBL having Arg72 is not due to a lack of this favorable interaction, but rather arises from UBA3's Arg190 acting as a negative gate. Thus, parallel residues from different UBL pathways can utilize distinct mechanisms to dictate interaction selectivity, and specificity can be amplified by barriers that prevent binding to components of different conjugation cascades.
Figures
Figure 1
Sequence conservation at a UBL’s residue 72 and E1 residues corresponding to UBA3’s 190. (A) Sequence alignment of the C-terminal tail region of NEDD8 and ubiquitin (UBIQ) from the following organisms: Hs, human; Mm, M. musculus; AT, A. thaliana; Ce, C. elegans; Dm, D. melanogaster. NEDD8 and ubiquitin residue 72 are highlighted. (B) Sequence alignment of the E1 region containing Arg190 (highlighted) from the UBA3 subunit of NEDD8’s E1 (E1 NEDD8) and the corresponding Gln from the UBA1 ubiquitin E1 (E1 UBIQ). (C) Nomenclature and sequences for NEDD8, ubiquitin, and E1 NEDD8 (APPBP1-UBA3) mutants.
Figure 2
Surface plasmon resonance analysis of APPBP1-UBA3 binding to UBLs. Representative sensorgrams (left) and binding curves (right) from surface plasmon resonance interaction assays, performed as described in , for (A) GST-APPBP1-UBA3Arg190 (wt), (B) GST-APPBP1-UBA3Arg190Gln, and (C) GST-APPBP1-UBA3Arg190Ala with NEDD8Ala72 (wt), NEDD8Ala72Arg, NEDD8Ala72Gln, and NEDD8Ala72Lys, as indicated. (D) Representative sensorgrams (left) and binding curves (right) for ubiquitin binding to GST-APPBP1-UBA3Arg190 (wt), GST-APPBP1-UBA3Arg190Gln, and GST-APPBP1-UBA3Arg190Ala, as indicated.
Figure 3
Altered E1 NEDD8 (APPBP1-UBA3)-E2 (Ubc12) transthiolation specificity for UBA3 Arg190 mutants. (A) Time-course of forming the Ubc12−NEDD8 thioester complexes with 100 nM wild-type and indicated mutants of APPBP1-UBA3, 4 μM wild-type and indicated mutants of NEDD8, and 3 μM Ubc12. Reactions were stopped at the indicated times, products were separated by SDS−PAGE and detected by Western blotting with anti-NEDD8 antibodies. (B) Western blots of reactions performed as in panel A, except using the indicated His-NEDD8 variants and His-ubiquitin, and probed with anti-His-tag antibodies.
Figure 4
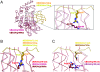
Structural basis for UBA3’s Arg190s negative selectivity against a UBL’s Arg72. Superimposition of wild-type (21) and mutant APPBP1-UBA3-NEDD8 structures was performed using least-squares fitting over all atoms in O (33). UBA3’s residue 190 is shown in various shades of red; NEDD8’s residue 72 in yellow; nitrogen, blue; and oxygen, light red. (A) Overall superimposition of APPBP1-UBA3Arg190Gln (rose)-NEDD8Ala72Arg (melon, “ubiquitinized”) and APPBP1-UBA3Arg190Ala (maroon)-NEDD8Ala72Arg (chartreuse, “wild-type-opposite”) complexes, with close-up view around the NEDD8 mutant’s Arg72 and UBA3’s residue 190. (B) Close-up view showing NEDD8 mutant Arg72 from APPBP1-UBA3Arg190Gln-NEDD8Ala72Arg (melon) and from APPBP1-UBA3Arg190Ala-NEDD8Ala72Arg (chartreuse), and Arg190 (red) from wild-type APPBP1-UBA3-NEDD8. (C) Close-up view showing Arg190 (violet) and NEDD8 mutant Gln72 from APPBP1-UBA3Arg190 (wt)-NEDD8Ala72Gln, and Arg190 (red) from wild-type APPBP1-UBA3-NEDD8.
Figure 5
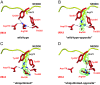
Differential APPBP1-UBA3 interactions with NEDD8 for UBA3 residue 190 and NEDD8 residue 72 mutants. Stick-representation close-up views, with UBA3 colored red and NEDD8 colored yellow, nitrogen blue, oxygen light-red, and hydrogen bonds shown as dashed lines for (A) APPBP1-UBA3-NEDD8 (wild-type), (B) APPBP1-UBA3Arg190Ala-NEDD8Ala72Arg (“wild-type-opposite”), (C) APPBP1-UBA3Arg190Gln-NEDD8Ala72Arg (“ubiquitinized”), and (D) APPBP1-UBA3Arg190 (wt)-NEDD8Ala72Gln (“ubiquitinized-opposite”) complexes. Simulated annealing omit Fo−Fc electron density maps are shown in green mesh, contoured at 3σ over UBA3’s residue 190 and NEDD8’s residue 72 in panels B−D. The maps were calculated using the program CNS (32), after simulated annealing at 2000 K omitting both UBA3’s residue 190 and NEDD8’s residue 72.
Similar articles
- The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1.
Walden H, Podgorski MS, Huang DT, Miller DW, Howard RJ, Minor DL Jr, Holton JM, Schulman BA. Walden H, et al. Mol Cell. 2003 Dec;12(6):1427-37. doi: 10.1016/s1097-2765(03)00452-0. Mol Cell. 2003. PMID: 14690597 - Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8's E1.
Huang DT, Paydar A, Zhuang M, Waddell MB, Holton JM, Schulman BA. Huang DT, et al. Mol Cell. 2005 Feb 4;17(3):341-50. doi: 10.1016/j.molcel.2004.12.020. Mol Cell. 2005. PMID: 15694336 - Basis for a ubiquitin-like protein thioester switch toggling E1-E2 affinity.
Huang DT, Hunt HW, Zhuang M, Ohi MD, Holton JM, Schulman BA. Huang DT, et al. Nature. 2007 Jan 25;445(7126):394-8. doi: 10.1038/nature05490. Epub 2007 Jan 14. Nature. 2007. PMID: 17220875 Free PMC article. - Chemical Tools for Probing the Ub/Ubl Conjugation Cascades.
Kochańczyk T, Fishman M, Lima CD. Kochańczyk T, et al. Chembiochem. 2025 Jan 2;26(1):e202400659. doi: 10.1002/cbic.202400659. Epub 2024 Nov 6. Chembiochem. 2025. PMID: 39313481 Free PMC article. Review. - Structural and functional insights to ubiquitin-like protein conjugation.
Streich FC Jr, Lima CD. Streich FC Jr, et al. Annu Rev Biophys. 2014;43:357-79. doi: 10.1146/annurev-biophys-051013-022958. Annu Rev Biophys. 2014. PMID: 24773014 Free PMC article. Review.
Cited by
- Inhibiting Neddylation with MLN4924 Suppresses Growth and Delays Multicellular Development in Dictyostelium discoideum.
Huber RJ, Kim WD, Mathavarajah S. Huber RJ, et al. Biomolecules. 2021 Mar 23;11(3):482. doi: 10.3390/biom11030482. Biomolecules. 2021. PMID: 33807046 Free PMC article. - The necessity of NEDD8/Rub1 for vitality and its association with mitochondria-derived oxidative stress.
Pick E. Pick E. Redox Biol. 2020 Oct;37:101765. doi: 10.1016/j.redox.2020.101765. Epub 2020 Oct 20. Redox Biol. 2020. PMID: 33099217 Free PMC article. Review. - Targeting NEDD8-activating enzyme for cancer therapy: developments, clinical trials, challenges and future research directions.
Fu DJ, Wang T. Fu DJ, et al. J Hematol Oncol. 2023 Jul 31;16(1):87. doi: 10.1186/s13045-023-01485-7. J Hematol Oncol. 2023. PMID: 37525282 Free PMC article. Review. - Neddylation of protein, a new strategy of protein post-translational modification for targeted treatment of central nervous system diseases.
Wu Q, Geng Z, Lu J, Wang S, Yu Z, Wang S, Ren X, Guan S, Liu T, Zhu C. Wu Q, et al. Front Neurosci. 2024 Nov 5;18:1467562. doi: 10.3389/fnins.2024.1467562. eCollection 2024. Front Neurosci. 2024. PMID: 39564524 Free PMC article. Review. - Phage display to identify Nedd8-mimicking peptides as inhibitors of the Nedd8 transfer cascade.
Zhao B, Zhang K, Villhauer EB, Bhuripanyo K, Kiyokawa H, Schindelin H, Yin J. Zhao B, et al. Chembiochem. 2013 Jul 22;14(11):1323-30. doi: 10.1002/cbic.201300234. Epub 2013 Jul 3. Chembiochem. 2013. PMID: 23824602 Free PMC article.
References
- Kerscher O.; Felberbaum R.; Hochstrasser M. (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22, 159–180. - PubMed
- Pickart C. M.; Fushman D. (2004) Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 8, 610–616. - PubMed
- Hochstrasser M. (2000) Evolution and function of ubiquitin-like protein-conjugation systems. Nature Cell Biol. 2, E153–157. - PubMed
- Pozo J. C.; Timpte C.; Tan S.; Callis J.; Estelle M. (1998) The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science 280, 1760–1763. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 CA071907/CA/NCI NIH HHS/United States
- 5P01CA0719075/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 GM077053/GM/NIGMS NIH HHS/United States
- P30CA021765/CA/NCI NIH HHS/United States
- R01GM077053/GM/NIGMS NIH HHS/United States
- R01 GM069530/GM/NIGMS NIH HHS/United States
- P30 CA021765/CA/NCI NIH HHS/United States
- R01GM069530/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous