Molecular architecture of native HIV-1 gp120 trimers - PubMed (original) (raw)
. 2008 Sep 4;455(7209):109-13.
doi: 10.1038/nature07159. Epub 2008 Jul 30.
Affiliations
- PMID: 18668044
- PMCID: PMC2610422
- DOI: 10.1038/nature07159
Molecular architecture of native HIV-1 gp120 trimers
Jun Liu et al. Nature. 2008.
Abstract
The envelope glycoproteins (Env) of human and simian immunodeficiency viruses (HIV and SIV, respectively) mediate virus binding to the cell surface receptor CD4 on target cells to initiate infection. Env is a heterodimer of a transmembrane glycoprotein (gp41) and a surface glycoprotein (gp120), and forms trimers on the surface of the viral membrane. Using cryo-electron tomography combined with three-dimensional image classification and averaging, we report the three-dimensional structures of trimeric Env displayed on native HIV-1 in the unliganded state, in complex with the broadly neutralizing antibody b12 and in a ternary complex with CD4 and the 17b antibody. By fitting the known crystal structures of the monomeric gp120 core in the b12- and CD4/17b-bound conformations into the density maps derived by electron tomography, we derive molecular models for the native HIV-1 gp120 trimer in unliganded and CD4-bound states. We demonstrate that CD4 binding results in a major reorganization of the Env trimer, causing an outward rotation and displacement of each gp120 monomer. This appears to be coupled with a rearrangement of the gp41 region along the central axis of the trimer, leading to closer contact between the viral and target cell membranes. Our findings elucidate the structure and conformational changes of trimeric HIV-1 gp120 relevant to antibody neutralization and attachment to target cells.
Figures
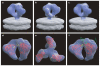
Figure 1. Averaged 3D structure of the HIV-1 spike in complex with b12-Fab
a, Perspective view of the surface of the density map shown at two thresholds; one to include the entire spike (outer), and another to highlight the Fab and gp120 components (inner). b—d, Front (b, c) and top (d) views of the map fitted with X-ray coordinates of the complex of the Fab fragment of b12 (cyan) with gp120 (red, PDB ID, 2NY7); only gp120 coordinates are shown in c, which is at the inner threshold. Likely locations of the V1/V2 loop and gp41 regions are indicated by asterisks in d and the white arrow in b, respectively. The stumps of the V1/V2 and V3 loop regions are shown in yellow and green, respectively.
Figure 2. Averaged 3D structure of the trimeric glycoprotein spike on native HIV-1
a, b Perspective/front views of the surface of the density map; the white arrow in b points to the likely location of gp41 in the map. c, Same view as in a but shown using two thresholds to illustrate both the overall shape (outer), and the contribution of the gp120 containing regions (inner). d, e, Front and top views of the map with coordinates for the gp120 core derived from the complex with b12 (PDB ID, 2NY7). f, Front view of the map fitted with coordinates for the gp120 core derived from the complex with X5 (PDB ID, 2B4C). The gp120 core is shown in red, and the regions of the V1/ V2 loop and V3 loop included in the coordinates are shown in yellow and green, respectively.
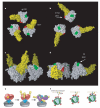
Figure 3. 3D structure of the HIV-1 spike in complex with CD4 and 17b-Fab
a, b Front and top views showing the X-ray coordinates of the ternary complex (PDB ID, 1GC1) of the gp120 core (red) with CD4 (yellow) and the Fab fragment of 17b (cyan) fitted into the map as a rigid body using automated fitting procedures. The arrow in a points to the likely location of the V1/V2 loop region, which was partially deleted in the construct used to obtain crystals of the ternary complex. c—e, Top views showing superposition of the X-ray coordinates for the gp120 trimer derived from the maps for the unliganded (white), b12-bound (cyan) and CD4/17b-bound (yellow) states of the trimeric spike, with locations of the V1/V2 stem regions indicated in red.
Figure 4. Description of the conformational change in the gp120 trimer induced by CD4 binding
a—d Model for the conformational change from the unliganded (a, c) to the CD4-bound state (b, d) shown as top (a, b) and front (c, d) views. The gp120 core, CD4, V1/V2 and V3 stems are shown in white, yellow, red and green colours, respectively. e, Schematic description of the gp41 (blue) and gp120 (red/purple) regions of the trimeric spike and the conformational changes that occur upon CD4 binding. The yellow patch near the apex marks the location of the CD4 binding site in the unliganded spike and the green patch at the apex marks the location of the V3 loop region in the spike after CD4 binding. f, Schematic view of the consequence of the CD4-induced conformational changes for viral attachment to the target cell and interaction with chemokine receptors (green at top). Colours in f have same meaning as in e.
Similar articles
- Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures.
Harris A, Borgnia MJ, Shi D, Bartesaghi A, He H, Pejchal R, Kang YK, Depetris R, Marozsan AJ, Sanders RW, Klasse PJ, Milne JL, Wilson IA, Olson WC, Moore JP, Subramaniam S. Harris A, et al. Proc Natl Acad Sci U S A. 2011 Jul 12;108(28):11440-5. doi: 10.1073/pnas.1101414108. Epub 2011 Jun 27. Proc Natl Acad Sci U S A. 2011. PMID: 21709254 Free PMC article. - Three-dimensional structures of soluble CD4-bound states of trimeric simian immunodeficiency virus envelope glycoproteins determined by using cryo-electron tomography.
White TA, Bartesaghi A, Borgnia MJ, de la Cruz MJ, Nandwani R, Hoxie JA, Bess JW, Lifson JD, Milne JL, Subramaniam S. White TA, et al. J Virol. 2011 Dec;85(23):12114-23. doi: 10.1128/JVI.05297-11. Epub 2011 Sep 21. J Virol. 2011. PMID: 21937655 Free PMC article. - Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop.
Wang H, Cohen AA, Galimidi RP, Gristick HB, Jensen GJ, Bjorkman PJ. Wang H, et al. Proc Natl Acad Sci U S A. 2016 Nov 15;113(46):E7151-E7158. doi: 10.1073/pnas.1615939113. Epub 2016 Oct 31. Proc Natl Acad Sci U S A. 2016. PMID: 27799557 Free PMC article. - Quaternary Interaction of the HIV-1 Envelope Trimer with CD4 and Neutralizing Antibodies.
Liu Q, Zhang P, Lusso P. Liu Q, et al. Viruses. 2021 Jul 20;13(7):1405. doi: 10.3390/v13071405. Viruses. 2021. PMID: 34372611 Free PMC article. Review. - HIV gp120: double lock strategy foils host defences.
Sattentau QJ. Sattentau QJ. Structure. 1998 Aug 15;6(8):945-9. doi: 10.1016/s0969-2126(98)00096-3. Structure. 1998. PMID: 9739096 Review.
Cited by
- Non-averaged single-molecule tertiary structures reveal RNA self-folding through individual-particle cryo-electron tomography.
Liu J, McRae EKS, Zhang M, Geary C, Andersen ES, Ren G. Liu J, et al. Nat Commun. 2024 Oct 21;15(1):9084. doi: 10.1038/s41467-024-52914-1. Nat Commun. 2024. PMID: 39433544 Free PMC article. - The asymmetric opening of HIV-1 Env by a potent CD4 mimetic enables anti-coreceptor binding site antibodies to mediate ADCC.
Richard J, Grunst MW, Niu L, Díaz-Salinas MA, Tolbert WD, Marchitto L, Zhou F, Bourassa C, Yang D, Chiu TJ, Chen HC, Benlarbi M; Guillaume-Beaudoin-Buissières; Gottumukkala S, Li W, Dionne K, Bélanger É, Chatterjee D, Medjahed H, Hendrickson WA, Sodroski J, Lang ZC, Morton AJ, Huang RK, Matthies D, Smith AB 3rd, Mothes W, Munro JB, Pazgier M, Finzi A. Richard J, et al. bioRxiv [Preprint]. 2024 Aug 27:2024.08.27.609961. doi: 10.1101/2024.08.27.609961. bioRxiv. 2024. PMID: 39253431 Free PMC article. Preprint. - Conformations of membrane human immunodeficiency virus (HIV-1) envelope glycoproteins solubilized in Amphipol A18 lipid-nanodiscs.
Zhang S, Anang S, Zhang Z, Nguyen HT, Ding H, Kappes JC, Sodroski J. Zhang S, et al. J Virol. 2024 Oct 22;98(10):e0063124. doi: 10.1128/jvi.00631-24. Epub 2024 Sep 9. J Virol. 2024. PMID: 39248459 - Conformational flexibility of HIV-1 envelope glycoproteins modulates transmitted/founder sensitivity to broadly neutralizing antibodies.
Parthasarathy D, Pothula KR, Ratnapriya S, Cervera Benet H, Parsons R, Huang X, Sammour S, Janowska K, Harris M, Sodroski J, Acharya P, Herschhorn A. Parthasarathy D, et al. Nat Commun. 2024 Aug 26;15(1):7334. doi: 10.1038/s41467-024-51656-4. Nat Commun. 2024. PMID: 39187497 Free PMC article. - Cryo-electron microscopy in the study of virus entry and infection.
Dutta M, Acharya P. Dutta M, et al. Front Mol Biosci. 2024 Jul 24;11:1429180. doi: 10.3389/fmolb.2024.1429180. eCollection 2024. Front Mol Biosci. 2024. PMID: 39114367 Free PMC article. Review.
References
- Dalgleish AG, et al. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. - PubMed
- Burton DR, et al. HIV vaccine design and the neutralizing antibody problem. Nature Immunol. 2004;5:233–236. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials