Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena - PubMed (original) (raw)
Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena
Lucia Aronica et al. Genes Dev. 2008.
Abstract
Tetrahymena eliminates micronuclear-limited sequences from the developing macronucleus during sexual reproduction. Homology between the sequences to be eliminated and approximately 28-nucleotide small RNAs (scnRNAs) associated with an Argonaute family protein Twi1p likely underlies this elimination process. However, the mechanism by which Twi1p-scnRNA complexes identify micronuclear-limited sequences is not well understood. We show that a Twi1p-associated putative RNA helicase Ema1p is required for the interaction between Twi1p and chromatin. This requirement explains the phenotypes of EMA1 KO strains, including loss of selective down-regulation of scnRNAs homologous to macronuclear-destined sequences, loss of H3K9 and K27 methylation in the developing new macronucleus, and failure to eliminate DNA. We further demonstrate that Twi1p interacts with noncoding transcripts derived from parental and developing macronuclei and this interaction is greatly reduced in the absence of Ema1p. We propose that Ema1p functions in DNA elimination by stimulating base-pairing interactions between scnRNAs and noncoding transcripts in both parental and developing new macronuclei.
Figures
Figure 1.
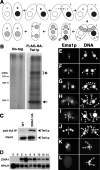
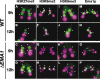
Identification and characterization of EMA1/Ema1p. (A) Conjugation. In the sexual process of conjugation, two cells mate (1); the Mic undergoes meiosis to form two haploid pronuclei, one of which is reciprocally exchanged between the two conjugating cells (2); the migratory and stationary pronuclei then fuse to create a zygotic nucleus (3); the zygotic nucleus divides mitotically twice to produce the next generation of new Macs and Mics (4); paired cells separate and one of the two new Mics and the parental Mac are destroyed (5); and cells resume vegetative growth with Mic mitosis, followed by cytokinesis (6). (B) Copurification of Ema1p with Flag-HA-Twi1p. Two wild-type strains (No-tag) or two Flag-HA-TWI1 strains were mated and were harvested at 9 h post-mixing. Flag-HA-Twi1p-containing complexes were enriched by gel filtration and immuno-affinity purified first with anti-Flag antibody and then with anti-HA antibody. The purified proteins were separated by SDS-PAGE and analyzed by silver staining. The arrow and the asterisk indicate the positions of Flag-HA-Twi1p and the Twi1p-associated proteins identified by mass spectrometry analysis, respectively. (C) Twi1p coimmunoprecipitates with Ema1p. An EMA1_-HA strain and a wild-type strain were mated and lysed at 6 h post-mixing. (Right) The Ema1p-HA-containing complex was pulled down with an anti-HA antibody. (Left) As a control, two wild-type strains were crossed and processed similarly. Coimmunoprecipitated proteins (anti-HA IP) or total proteins (Input) used for immunoprecipitation were analyzed on Western blot using anti-Twi1p antiserum. (D) Expression of EMA1 mRNA. Total RNAs from exponentially growing (E), starved (S), and conjugating (2, 4, 6, 8, 10, 12, and 14 h post-mixing) wild-type cells were analyzed by Northern hybridization. RPL21 was used as a loading control. (E–L) Localization of Ema1p. Mating pairs of wild-type cells in the early (E) and late (F) premeiosis, pronuclear exchange (G), Mac Anlagen (H,I), Nuclear Alignment (J), Pair Separation (K), and Mic Elimination (L) stages were processed for immunostaining. See Supplemental Figure S1 for the conjugation stages. Ema1p was localized using anti-Ema1p antiserum (left) and DNA was stained by DAPI (right). Arrows, arrowheads, and arrowheads marked with “An” indicate Macs, Mics, and developing new Macs, respectively. The staining detected at the junction of cells (double arrowheads) was also observed in Δ_EMA1 strains (see Fig. 7O,P) and thus represents cross-reaction of the antiserum with other proteins.
Figure 2.
Characterization of Δ_EMA1_ cells. (A) EMA1 locus and KO construct. A part of the EMA1 coding sequence, including the conserved helicase domains (shown in gray) was replaced by the drug-resistance marker neo3. Upon transformation, the KO construct was introduced into the EMA1 locus by homologous recombination. (B) Southern hybridization of Δ_EMA1_ strains. Total DNA isolated from wild-type (WT) or Δ_EMA1_ strains was digested with NdeI (N in A), and the blot was hybridized with the probe shown in A. Positions of the bands for wild-type and KO loci are indicated with arrowheads. (C) Ema1p expression in Δ_EMA1_ strains. (Top panel) Ema1p expression in the wild-type (W) and Δ_EMA1_ (Δ) strains in starved (S) and mating (4, 8, and 12 h post-mixing) cells was analyzed by Western blot using anti-Ema1p antiserum. (Bottom panel) For a loading control, the amount of α-tubulin was analyzed. (D) Developmental profiles of conjugation in wild-type and Δ_EMA1_ strains. Conjugation stage wild-type (CU427 × CU428) and Δ_EMA1_ [(7)-3-1 X (8)-1-1] cells were observed by DAPI staining. The stages categorized were single unmated cells (S), premeiosis (E1), meiosis (E2), prezygotic (M1), post-zygotic (M2), Mac development (L1), Pair Separation (2 Mics) (L2), and Mic elimination (L3). See Supplemental Figure S1 for the developmental stages. (E) Δ_EMA1_ causes arrest at Pair Separation stage. At 36 h post-mixing, the progeny of wild-type (CU427 and CU428) or Δ_EMA1_ cells were fixed and nuclei were observed by DAPI staining. The stages categorized were the same as in D except stages E1∼L1 were combined. (F) Δ_EMA1_ cells fail to produce viable progeny. At 6∼8 h post-mixing, single mating pairs were placed into drops of medium and incubated for ∼60 h at 30°C. Completion of conjugation was confirmed by testing for expression of the marker specific for newly developed Macs.
Figure 3.
DNA elimination in the progeny of ΔEMA1 cells. (Left) Schematic drawings of the IES elimination assays. Solid horizontal lines indicate MDSs and the open boxes indicate IESs. The M and R elements are ∼2.5 kb apart on the same Mic chromosome. Four primers (arrows) that flank each IES were used for nested PCR. For the assays of Cal and Tlr-1 elements, primers complementary to IESs were also used for the same PCR. (Right) Results of the IES elimination assays. The sizes of the unprocessed (Mic form) and the processed (Mac form) products are marked by arrowheads with “i” and “a”, respectively. (m) Molecular weight marker.
Figure 4.
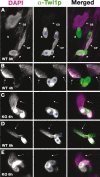
Association of Twi1p with chromatin in Δ_EMA1_ cells. (A–E) Cells with partially spread chromatin from wild-type (A,B,D) or Δ_EMA1_ (C,E) strains were fixed at the indicated time points post-mixing (mating). The localization of Twi1p (middle column; green in the right column) was analyzed by immunostaining using an anti-Twi1p antibody. DNA was counterstained with DAPI (left column; purple in the right column). (S) Single, nonmating cell; (P) paired mating cells; (CS) chromatin from a single cell; (CP) chromatin from paired cells. Arrows in B–E indicate chromatin from mating cells. In this experiment, the cell cortex (blue asterisks) has background staining for anti-Twi1p.
Figure 5.
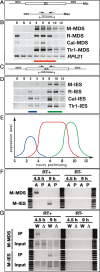
Three types ncRNA. (A–D) Exponentially growing (E), starved (S), and conjugating (2, 4, 6, 8, 10, and 12 h post-mixing) wild-type cells were analyzed by RT–PCR. (A) Schematic drawing of the analysis of ncRNA transcription from the parental Mac. Sets of primers (arrows) that flank four different IESs were used to amplify the cDNAs. (B) Results of RT–PCR for Mac loci flanking the indicated elements. The red horizontal bar indicates stages in which the ncRNAs are up-regulated. Constitutively expressed RPL21 mRNA was amplified as the positive control for the RT reaction. (C) Schematic drawing of ncRNA analysis from IESs. Sets of primers (arrows) in four different IESs were used to amplify the cDNAs. (D) The results of RT–PCR for the indicated IES elements. Blue and green horizontal bars indicate stages in which the ncRNAs are up-regulated. (E) Schematic summary of ncRNA transcription analyses. Relative expression levels (vertical axis) and expression timing (horizontal axis) of the ncRNAs from the Mic (blue), the parental Mac (red), and the developing Mac (green) are represented. (F) Coimmunoprecipitation of ncRNAs with Twi1p. (Lanes labeled as A) Conjugating wild-type strains at 4.5 or 9 h post-mixing were lysed and the Twi1p complex was immunoprecipitated with anti-Twi1p antibody. Coimmunoprecipitated RNA was analyzed by RT–PCR (RT+) to detect M-MDS (from parental Mac) or M-IES (from developing new Mac) transcripts as above. As a control, a similar experiment was performed with preimmune serum (lanes labeled as P). In parallel, a similar experiment was performed without the reverse transcription reaction (RT−). (G) Coimmunoprecipitation assay of ncRNAs with Twi1p in the wild-type (W) or Δ_EMA1_ strains (Δ). Conjugating cells were lysed at 4.5 or 9 h post-mixing and the Twi1p complex was immunoprecipitated with anti-Twi1p antibody (IP). Coimmunoprecipitated RNA was analyzed by RT–PCR (RT+) to detect M-MDS or M-IES transcripts. RNA was also extracted from part of the lysate and analyzed by RT–PCR (Input). As a negative control, a similar experiment was performed without the reverse transcription reaction (RT−).
Figure 6.
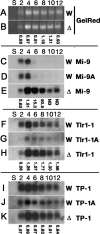
Expression of scnRNAs in Δ_EMA1_ cells. Total RNA was extracted from starved (S) or conjugating (2, 4, 6, 8, 10, and 12 h post-mixing) wild-type (W) or Δ_EMA1_ (Δ) strains and separated in sequencing gels. (A,B) Bulk scnRNAs (∼28 nt) were visualized by staining the gels with GelRed. (C–K) Blots were hybridized with the probes indicated (right). Quantitation of the signals are shown as ratios of the signals (Δ_EMA1_/wild-type) obtained from gel staining and Northern blots.
Figure 7.
H3K9/K27me in Δ_EMA1_ cells. Wild-type (WT, top) and Δ_EMA1_ (bottom) conjugating cells at 9 or 12 h post-mixing were processed for immunofluorescent staining (green) using anti-H3K27me3 (A–D), anti-H3K9me2 (E–H), anti-H3K9me3 (I–L), or anti-Ema1p (M–P) Abs. DNA was stained with DAPI (purple). Arrowheads indicate developing new Macs.
Figure 8.
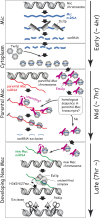
A refined scnRNA model. The sequentially occurring events are drawn from top to bottom. The approximate stages when the events occur are shown on the right with arrows. See the text for details.
Similar articles
- Two GW repeat proteins interact with Tetrahymena thermophila argonaute and promote genome rearrangement.
Bednenko J, Noto T, DeSouza LV, Siu KW, Pearlman RE, Mochizuki K, Gorovsky MA. Bednenko J, et al. Mol Cell Biol. 2009 Sep;29(18):5020-30. doi: 10.1128/MCB.00076-09. Epub 2009 Jul 13. Mol Cell Biol. 2009. PMID: 19596782 Free PMC article. - Biased transcription and selective degradation of small RNAs shape the pattern of DNA elimination in Tetrahymena.
Schoeberl UE, Kurth HM, Noto T, Mochizuki K. Schoeberl UE, et al. Genes Dev. 2012 Aug 1;26(15):1729-42. doi: 10.1101/gad.196493.112. Genes Dev. 2012. PMID: 22855833 Free PMC article. - Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena.
Liu Y, Mochizuki K, Gorovsky MA. Liu Y, et al. Proc Natl Acad Sci U S A. 2004 Feb 10;101(6):1679-84. doi: 10.1073/pnas.0305421101. Epub 2004 Jan 30. Proc Natl Acad Sci U S A. 2004. PMID: 14755052 Free PMC article. - Programmed DNA elimination in Tetrahymena: a small RNA-mediated genome surveillance mechanism.
Kataoka K, Mochizuki K. Kataoka K, et al. Adv Exp Med Biol. 2011;722:156-73. doi: 10.1007/978-1-4614-0332-6_10. Adv Exp Med Biol. 2011. PMID: 21915788 Free PMC article. Review. - Small RNAs in genome rearrangement in Tetrahymena.
Mochizuki K, Gorovsky MA. Mochizuki K, et al. Curr Opin Genet Dev. 2004 Apr;14(2):181-7. doi: 10.1016/j.gde.2004.01.004. Curr Opin Genet Dev. 2004. PMID: 15196465 Review.
Cited by
- Functional Proteomics of Nuclear Proteins in Tetrahymena thermophila: A Review.
Saettone A, Nabeel-Shah S, Garg J, Lambert JP, Pearlman RE, Fillingham J. Saettone A, et al. Genes (Basel). 2019 May 1;10(5):333. doi: 10.3390/genes10050333. Genes (Basel). 2019. PMID: 31052454 Free PMC article. Review. - RNA-Mediated Epigenetic Programming of Genome Rearrangements.
Nowacki M, Shetty K, Landweber LF. Nowacki M, et al. Annu Rev Genomics Hum Genet. 2011;12:367-89. doi: 10.1146/annurev-genom-082410-101420. Annu Rev Genomics Hum Genet. 2011. PMID: 21801022 Free PMC article. Review. - Whats, hows and whys of programmed DNA elimination in Tetrahymena.
Noto T, Mochizuki K. Noto T, et al. Open Biol. 2017 Oct;7(10):170172. doi: 10.1098/rsob.170172. Open Biol. 2017. PMID: 29021213 Free PMC article. Review. - Programmed chromosome fragmentation in ciliated protozoa: multiple means to chromosome ends.
Bétermier M, Klobutcher LA, Orias E. Bétermier M, et al. Microbiol Mol Biol Rev. 2023 Dec 20;87(4):e0018422. doi: 10.1128/mmbr.00184-22. Epub 2023 Nov 27. Microbiol Mol Biol Rev. 2023. PMID: 38009915 Free PMC article. Review. - Functional study of genes essential for autogamy and nuclear reorganization in Paramecium.
Nowak JK, Gromadka R, Juszczuk M, Jerka-Dziadosz M, Maliszewska K, Mucchielli MH, Gout JF, Arnaiz O, Agier N, Tang T, Aggerbeck LP, Cohen J, Delacroix H, Sperling L, Herbert CJ, Zagulski M, Bétermier M. Nowak JK, et al. Eukaryot Cell. 2011 Mar;10(3):363-72. doi: 10.1128/EC.00258-10. Epub 2011 Jan 21. Eukaryot Cell. 2011. PMID: 21257794 Free PMC article.
References
- Bühler M., Verdel A., Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials