Oncogenic mutations in GNAQ occur early in uveal melanoma - PubMed (original) (raw)
Oncogenic mutations in GNAQ occur early in uveal melanoma
Michael D Onken et al. Invest Ophthalmol Vis Sci. 2008 Dec.
Abstract
Purpose: Early/initiating oncogenic mutations have been identified for many cancers, but such mutations remain unidentified in uveal melanoma (UM). An extensive search for such mutations was undertaken, focusing on the RAF/MEK/ERK pathway, which is often the target of initiating mutations in other types of cancer.
Methods: DNA samples from primary UMs were analyzed for mutations in 24 potential oncogenes that affect the RAF/MEK/ERK pathway. For GNAQ, a stimulatory alpha(q) G-protein subunit which was recently found to be mutated in UMs, resequencing was expanded to include 67 primary UMs and 22 peripheral blood samples. GNAQ status was analyzed for association with clinical, pathologic, chromosomal, immunohistochemical, and transcriptional features.
Results: Activating mutations at codon 209 were identified in GNAQ in 33 (49%) of 67 primary UMs, including 2 (22%) of 9 iris melanomas and 31 (54%) of 58 posterior UMs. No mutations were found in the other 23 potential oncogenes. GNAQ mutations were not found in normal blood DNA samples. Consistent with GNAQ mutation being an early or initiating event, this mutation was not associated with any clinical, pathologic, or molecular features associated with late tumor progression.
Conclusions: GNAQ mutations occur in about half of UMs, representing the most common known oncogenic mutation in this cancer. The presence of this mutation in tumors at all stages of malignant progression suggests that it is an early event in UM. Mutations in this G-protein-coupled receptor provide new insights into UM pathogenesis and could lead to new therapeutic possibilities.
Figures
Figure 1
Regions of chromosomal gain identified by CGH. (A) CGHminer result for sixteen class 1 and twelve class 2 tumors. DNA gains (indicated by orange and red vertical bars) with respect to chromosomal position (horizontal lines) on chromosomes 6p, 8q and, to a lesser extent, 20p and 20q. The p-arms are depicted to the left, and the q-arms to the right of the centromeres (vertical purple bars). The percent of samples showing DNA gain is indicated by the scale at the bottom. (B) CGH tracing of chromosome 5, showing two peaks with a mean log2ratio ≥ 3 standard deviations above the mean for the chromosomal arm. The larger peak at 5q13.1 corresponded to the location of PIK3R1. The other smaller peak did not correspond to a coding region. (C) Pathways that affect RAF/MEK/ERK activation. Arrows indicate stimulatory interactions, and T-bars indicate inhibitory interactions. Abbreviations: RTK, receptor tyrosine kinase; GPCR, G-protein coupled receptor. Other abbreviations are official gene symbols. Notations of specific genes analyzed in this study: 1Ras superfamily of small GTPases: DIRAS2, REM1, GEM, RAB2A, RAB22A, RAB23 (HRAS, KRAS and NRAS were previously analyzed); 2PAK7; 3ARAF, RAF1 and RASIP1 (BRAF was previously analyzed); 4PTK2 and PTK6; 5PIK3R1 regulatory subunit; 6MAPK13 and MAPK14; 7GNAQ; 8GRM1. Red shapes indicate genes that were re-sequenced in this study.
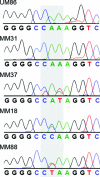
Figure 2
Representative sequence tracings for GNAQ surrounding codon 209 (shaded). UM86, normal uveal melanocyte sample; MM31, uveal melanoma with wildtype sequence (CAA); MM37, MM18 and MM88, uveal melanomas with the three mutant sequences, as indicated.
Similar articles
- Prognostic impact of chromosomal aberrations and GNAQ, GNA11 and BAP1 mutations in uveal melanoma.
Staby KM, Gravdal K, Mørk SJ, Heegaard S, Vintermyr OK, Krohn J. Staby KM, et al. Acta Ophthalmol. 2018 Feb;96(1):31-38. doi: 10.1111/aos.13452. Epub 2017 Apr 26. Acta Ophthalmol. 2018. PMID: 28444874 - Identification of unique MEK-dependent genes in GNAQ mutant uveal melanoma involved in cell growth, tumor cell invasion, and MEK resistance.
Ambrosini G, Pratilas CA, Qin LX, Tadi M, Surriga O, Carvajal RD, Schwartz GK. Ambrosini G, et al. Clin Cancer Res. 2012 Jul 1;18(13):3552-61. doi: 10.1158/1078-0432.CCR-11-3086. Epub 2012 May 1. Clin Cancer Res. 2012. PMID: 22550165 Free PMC article. - Lack of oncogenic GNAQ mutations in melanocytic lesions of the conjunctiva as compared to uveal melanoma.
Dratviman-Storobinsky O, Cohen Y, Frenkel S, Pe'er J, Goldenberg-Cohen N. Dratviman-Storobinsky O, et al. Invest Ophthalmol Vis Sci. 2010 Dec;51(12):6180-2. doi: 10.1167/iovs.10-5677. Epub 2010 Jul 14. Invest Ophthalmol Vis Sci. 2010. PMID: 20631239 - What hope for the future? GNAQ and uveal melanoma.
Sisley K, Doherty R, Cross NA. Sisley K, et al. Br J Ophthalmol. 2011 May;95(5):620-3. doi: 10.1136/bjo.2010.182097. Epub 2011 Mar 3. Br J Ophthalmol. 2011. PMID: 21378004 Review. - Research in practice: Therapeutic targeting of oncogenic GNAQ mutations in uveal melanoma.
Gaffal E. Gaffal E. J Dtsch Dermatol Ges. 2020 Nov;18(11):1245-1248. doi: 10.1111/ddg.14288. Epub 2020 Sep 21. J Dtsch Dermatol Ges. 2020. PMID: 32954611 Review.
Cited by
- Patient survival in uveal melanoma is not affected by oncogenic mutations in GNAQ and GNA11.
Koopmans AE, Vaarwater J, Paridaens D, Naus NC, Kilic E, de Klein A; Rotterdam Ocular Melanoma Study group. Koopmans AE, et al. Br J Cancer. 2013 Jul 23;109(2):493-6. doi: 10.1038/bjc.2013.299. Epub 2013 Jun 18. Br J Cancer. 2013. PMID: 23778528 Free PMC article. - Repression of genes involved in melanocyte differentiation in uveal melanoma.
Bergeron MA, Champagne S, Gaudreault M, Deschambeault A, Landreville S. Bergeron MA, et al. Mol Vis. 2012;18:1813-22. Epub 2012 Jul 4. Mol Vis. 2012. PMID: 22815634 Free PMC article. - MiR-181a-5p inhibits uveal melanoma development by targeting GNAQ and AKT3.
Wang R, Tahiri H, Yang C, Landreville S, Callejo S, Hardy P. Wang R, et al. Am J Cancer Res. 2023 Jan 15;13(1):293-306. eCollection 2023. Am J Cancer Res. 2023. PMID: 36777504 Free PMC article. - Mechanisms of resistance to imatinib mesylate in KIT-positive metastatic uveal melanoma.
Calipel A, Landreville S, De La Fouchardière A, Mascarelli F, Rivoire M, Penel N, Mouriaux F. Calipel A, et al. Clin Exp Metastasis. 2014 Jun;31(5):553-64. doi: 10.1007/s10585-014-9649-2. Epub 2014 Mar 21. Clin Exp Metastasis. 2014. PMID: 24652072 - Targeted therapy for melanoma: a primer.
Davies MA, Gershenwald JE. Davies MA, et al. Surg Oncol Clin N Am. 2011 Jan;20(1):165-80. doi: 10.1016/j.soc.2010.09.003. Surg Oncol Clin N Am. 2011. PMID: 21111965 Free PMC article. Review.
References
- Petrausch U, Martus P, Tonnies H, et al. Significance of gene expression analysis in uveal melanoma in comparison to standard risk factors for risk assessment of subsequent metastases. Eye. 2007 published online. - PubMed
- Tschentscher F, Husing J, Holter T, et al. Tumor classification based on gene expression profiling shows that uveal melanomas with and without monosomy 3 represent two distinct entities. Cancer Res. 2003;63:2578–2584. - PubMed
- Dahl C, Guldberg P. The genome and epigenome of malignant melanoma. APMIS. 2007;115:1161–1176. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 EY1316905/EY/NEI NIH HHS/United States
- P30 EY002687/EY/NEI NIH HHS/United States
- R01 EY013169/EY/NEI NIH HHS/United States
- R01 EY013169-05/EY/NEI NIH HHS/United States
- R01 CA125970/CA/NCI NIH HHS/United States
- P30 EY 02687/EY/NEI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous