Biochemical and structural insights of the early glycosylation steps in calicheamicin biosynthesis - PubMed (original) (raw)
Biochemical and structural insights of the early glycosylation steps in calicheamicin biosynthesis
Changsheng Zhang et al. Chem Biol. 2008.
Abstract
The enediyne antibiotic calicheamicin (CLM) gamma(1)(I) is a prominent antitumor agent that is targeted to DNA by a novel aryltetrasaccharide comprised of an aromatic unit and four unusual carbohydrates. Herein we report the heterologous expression and the biochemical characterization of the two "internal" glycosyltransferases CalG3 and CalG2 and the structural elucidation of an enediyne glycosyltransferase (CalG3). In conjunction with the previous characterization of the "external" CLM GTs CalG1 and CalG4, this study completes the functional assignment of all four CLM GTs, extends the utility of enediyne GT-catalyzed reaction reversibility, and presents conclusive evidence of a sequential glycosylation pathway in CLM biosynthesis. This work also reveals the common GT-B structural fold can now be extended to include enediyne GTs.
Figures
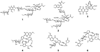
Fig. 1. Representative naturally occurring enediynes
ncluding the 10-membered enediynes calicheamcin γ1I (1), esperamicin (2), dynemicin (3) and 9-membered chromoprotein enediynes C-1027 (4), neocarzinostatin (5), and maduropeptin (6). The CLM aryltetrasaccharide is highlighted in blue.
Fig. 2. Preparation of CLM T0 (9) from CLM α3I (7)
(A) Schematic of the strategy – (a) refluxing in acetone, 65°C, 20 h; (b) incubated in 0.2% TFA, RT, 4 h. (B) HPLC analyses of the preparation - (i) the starting material 7; (ii) refluxed for 20 h; (iii) purified 8 from the reaction mixture of (ii); (iv) 8 incubated with 0.2% TFA at RT for 2 h; and (iv) 8 incubated with 0.2% TFA at RT for 4 h.
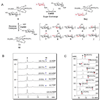
Fig. 3. CalG3-catalyzed reverse reaction and ‘sugar exchange’ reaction
(A) Schematic of the CalG3-catalyzed formation of 10 from 9 via reverse catalysis and the production of 10 novel CLM variants 9a–j via ‘sugar exchange’. (B) HPLC analyses of CalG3-catalyzed reverse reactions. In these reactions, 50 µM 9 was incubated with 7.5 µM CalG3 for 2 h at 30 °C in the presence of (i) 2 mM ADP; (ii) 2 mM GDP; (iii) 2 mM UDP; (iv) 2 mM CDP; (v) 2 mM TDP and (vi) no NDP. Percent conversions were indicated in the parentheses. (C) The production of 9a–j via CalG3-catalyzed ‘sugar exchange’. 50 µM 9 was incubated with 7.5 µM CalG3 at 30°C overnight in the presence of various TDP-sugars (300 µM, Fig. S4). Percent conversions were indicated in the parentheses.
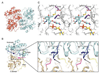
Fig. 4. Structure of CalG3
(A) A ribbon diagram of the CalG3 dimer with monomers color-coded in red and cyan. (B) The CalG3 monomer is formed by closely opposed N-terminal- (cyan) and the C-terminal-domains (khaki). These distinct domains are connected by a linker (yellow) and their interaction is stabilized by the C-terminal helix (green). The blue arrow indicates the putative catalytic loop, the magenta arrow points to a pyrophosphate-binding tetraglycine loop spanning residues 285–288. An ordered portion of a polyethylene glycol molecule (brown) has been found in the cavity formed by the N-domain. The inset highlights Cot-trace of CalG3 (cyan, yellow, khaki) in the active site with putative catalytic diad residues H11 and E115 highlighted. The gray Cα-trace is that of a docked model, which incorporates experimentally-observed conformational changes in the pyrophosphate binding loop (magenta) and modeled changes of the "catalytic loop" (blue, black arrow). (C) Manually docked model of the CalG3 with CLM T0 (carbon-cyan, oxygen-red, sulfur-yellow, nitrogen-blue) and a dinucleotide TDP (carbon-yellow, oxygen-red, nitrogen-blue, phosphorus-orange) in the active site.
Fig. 5. Differential reactions with 9 and 12 in the presence of CalG2 and CalG3
(i) 50 µM 9, 300 µM 12 in the presence of 7.5 µM CalG2 at 30 °C overnight; (ii) 50 µM 9, 300 µM 12 in the absence of enzymes at 30 °C overnight; (iii) 50 µM 9, 300 µM 12 in the presence of 7.5 µM CalG3 at 30 °C overnight.
Fig. 6. The proposed CLM glycosylation pathway
Similar articles
- Exploiting the reversibility of natural product glycosyltransferase-catalyzed reactions.
Zhang C, Griffith BR, Fu Q, Albermann C, Fu X, Lee IK, Li L, Thorson JS. Zhang C, et al. Science. 2006 Sep 1;313(5791):1291-4. doi: 10.1126/science.1130028. Science. 2006. PMID: 16946071 - Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Chang A, Singh S, Helmich KE, Goff RD, Bingman CA, Thorson JS, Phillips GN Jr. Chang A, et al. Proc Natl Acad Sci U S A. 2011 Oct 25;108(43):17649-54. doi: 10.1073/pnas.1108484108. Epub 2011 Oct 10. Proc Natl Acad Sci U S A. 2011. PMID: 21987796 Free PMC article. - Solution structure of the calicheamicin gamma 1I-DNA complex.
Kumar RA, Ikemoto N, Patel DJ. Kumar RA, et al. J Mol Biol. 1997 Jan 17;265(2):187-201. doi: 10.1006/jmbi.1996.0718. J Mol Biol. 1997. PMID: 9020982 - Glycosyltransferase structural biology and its role in the design of catalysts for glycosylation.
Chang A, Singh S, Phillips GN Jr, Thorson JS. Chang A, et al. Curr Opin Biotechnol. 2011 Dec;22(6):800-8. doi: 10.1016/j.copbio.2011.04.013. Epub 2011 May 16. Curr Opin Biotechnol. 2011. PMID: 21592771 Free PMC article. Review. - Glycosyltransferases: mechanisms and applications in natural product development.
Liang DM, Liu JH, Wu H, Wang BB, Zhu HJ, Qiao JJ. Liang DM, et al. Chem Soc Rev. 2015 Nov 21;44(22):8350-74. doi: 10.1039/c5cs00600g. Epub 2015 Sep 2. Chem Soc Rev. 2015. PMID: 26330279 Review.
Cited by
- Structural and functional characterization of CalS11, a TDP-rhamnose 3'-O-methyltransferase involved in calicheamicin biosynthesis.
Singh S, Chang A, Helmich KE, Bingman CA, Wrobel RL, Beebe ET, Makino S, Aceti DJ, Dyer K, Hura GL, Sunkara M, Morris AJ, Phillips GN Jr, Thorson JS. Singh S, et al. ACS Chem Biol. 2013 Jul 19;8(7):1632-9. doi: 10.1021/cb400068k. Epub 2013 May 23. ACS Chem Biol. 2013. PMID: 23662776 Free PMC article. - Modulation of deoxysugar transfer by the elloramycin glycosyltransferase ElmGT through site-directed mutagenesis.
Ramos A, Olano C, Braña AF, Méndez C, Salas JA. Ramos A, et al. J Bacteriol. 2009 Apr;191(8):2871-5. doi: 10.1128/JB.01747-08. Epub 2009 Feb 20. J Bacteriol. 2009. PMID: 19233921 Free PMC article. - OleD Loki as a Catalyst for Tertiary Amine and Hydroxamate Glycosylation.
Hughes RR, Shaaban KA, Zhang J, Cao H, Phillips GN Jr, Thorson JS. Hughes RR, et al. Chembiochem. 2017 Feb 16;18(4):363-367. doi: 10.1002/cbic.201600676. Epub 2017 Jan 9. Chembiochem. 2017. PMID: 28067448 Free PMC article. - Antibody-drug conjugates, cancer immunotherapy, and metronomic chemotherapy as novel approaches in cancer management.
Sarangi SC, Sopory P, Pattnaik SS, Reeta KH. Sarangi SC, et al. Indian J Pharmacol. 2020 Sep-Oct;52(5):402-413. doi: 10.4103/ijp.IJP_475_18. Indian J Pharmacol. 2020. PMID: 33283772 Free PMC article. Review. - Understanding substrate selectivity of human UDP-glucuronosyltransferases through QSAR modeling and analysis of homologous enzymes.
Dong D, Ako R, Hu M, Wu B. Dong D, et al. Xenobiotica. 2012 Aug;42(8):808-20. doi: 10.3109/00498254.2012.663515. Epub 2012 Mar 2. Xenobiotica. 2012. PMID: 22385482 Free PMC article. Review.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. - PubMed
- Ahlert J, Shepard E, Lomovskaya N, Zazopoulos E, Staffa A, Bachmann BO, Huang K, Fonstein L, Czisny A, Whitwam RE, et al. The calicheamicin gene cluster and its iterative type I enediyne PKS. Science. 2002;297:1173–1176. - PubMed
- Biggins JB, Onwueme KC, Thorson JS. Resistance to enediyne antitumor antibiotics by CalC self-sacrifice. Science. 2003;301:1537–1541. - PubMed
- Bililign T, Shepard EM, Ahlert J, Thorson JS. On the origin of deoxypentoses: evidence to support a glucose progenitor in the biosynthesis of calicheamicin. ChemBioChem. 2002;3:1143–1146. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U19 CA 113297/CA/NCI NIH HHS/United States
- U19 CA113297-030002/CA/NCI NIH HHS/United States
- R01 CA084374/CA/NCI NIH HHS/United States
- U19 CA113297-050002/CA/NCI NIH HHS/United States
- R01 CA084374-11/CA/NCI NIH HHS/United States
- CO 1020/CO/NCI NIH HHS/United States
- R01 CA084374-10/CA/NCI NIH HHS/United States
- GM 1104/GM/NIGMS NIH HHS/United States
- U19 CA113297/CA/NCI NIH HHS/United States
- R01 CA084374-12/CA/NCI NIH HHS/United States
- CA 84374/CA/NCI NIH HHS/United States
- U19 CA113297-040002/CA/NCI NIH HHS/United States
- R01 CA084374-09/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous