Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum - PubMed (original) (raw)
Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum
Elizabeth L Axe et al. J Cell Biol. 2008.
Abstract
Autophagy is the engulfment of cytosol and organelles by double-membrane vesicles termed autophagosomes. Autophagosome formation is known to require phosphatidylinositol 3-phosphate (PI(3)P) and occurs near the endoplasmic reticulum (ER), but the exact mechanisms are unknown. We show that double FYVE domain-containing protein 1, a PI(3)P-binding protein with unusual localization on ER and Golgi membranes, translocates in response to amino acid starvation to a punctate compartment partially colocalized with autophagosomal proteins. Translocation is dependent on Vps34 and beclin function. Other PI(3)P-binding probes targeted to the ER show the same starvation-induced translocation that is dependent on PI(3)P formation and recognition. Live imaging experiments show that this punctate compartment forms near Vps34-containing vesicles, is in dynamic equilibrium with the ER, and provides a membrane platform for accumulation of autophagosomal proteins, expansion of autophagosomal membranes, and emergence of fully formed autophagosomes. This PI(3)P-enriched compartment may be involved in autophagosome biogenesis. Its dynamic relationship with the ER is consistent with the idea that the ER may provide important components for autophagosome formation.
Figures
Figure 1.
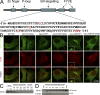
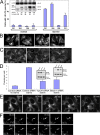
Identification and partial characterization of an ER-targeting domain within DFCP1. (A) Sequential truncations of DFCP1 identified an internal domain necessary and sufficient to specify localization in the ER. Residues in red and underlined are required for ER localization. (B) The GFP-[416–543] construct localized to the ER in HEK-293 cells, whereas mutants within this domain (shown here is a W543A construct) were cytosolic. Selected residues important in ER targeting were also mutagenized in the full-length protein as indicated. The full-length DFCP1 (myc-DFCP1) localized to the ER, whereas the DM (myc-dmDFCP1) had ER/Golgi localization (insets in myc-dmDFCP1 panels show colocalization with giantin). Mutagenesis of critical residues resulted in cytoplasmic redistribution (shown here as W/A mutant). All samples were costained with antibodies to calnexin (CLNX), an ER protein. (C) The WT or the W/A mutant of the 416–543 domain were purified as GST fusion proteins (input) and mixed with rat kidney microsomes. Bound material was recovered by centrifugation. Note that the WT-derived protein bound to microsomes, whereas the W/A mutant did not. (D) Microsomes as in C were treated on ice with the indicated units of trypsin before incubation with the GST-[416–543] domain and centrifugation. Note that β-COP, a peripheral protein found on microsomes, was almost completely digested by trypsin; under these conditions, binding of the DFCP1 fragment to microsomes was not changed.
Figure 2.
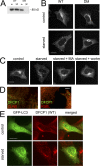
Starvation-induced and PI(3)P-dependent translocation of DFCP1 to punctate structures partially colocalizing with autophagosomes. (A) Lysates from cells expressing WT DFCP1 or the DM were incubated with Affigel beads coupled to PI(3)P; WT DFCP1 bound to PI(3)P, whereas the DM did not. in, 5% of input. (B) Stable HEK-293 cell lines expressing WT DFCP1 or the DM were left untreated or starved of amino acids for 90 min. Cells were then fixed and stained for DFCP1. Both WT and DM DFCP1 under normal conditions localized to the ER/Golgi; after starvation, only the WT translocated to punctate structures. (C) HEK-293 cells stably expressing WT DFCP1 were starved alone or in the presence of 3-methyladenine (MA) or wortmannin (wortm) as indicated before fixation and staining for DFCP1. (D) Starved HEK-293 cells expressing DFCP1 were costained with markers to the ER such as calnexin and KDEL. (E) HEK-293 cells stably expressing WT DFCP1 were transfected with GFP-MAP-LC3 and starved for 90 min as indicated. Note that in cells expressing WT DFCP1, both MAP-LC3 and DFCP1 translocated to punctate structures upon starvation, some of which colocalized. Bars: (A–C and E) 20 μm; (D) 0.5 μm.
Figure 3.
FYVE domain tethered peripherally to the ER translocates to starvation-induced punctate structures in a pathway dependent on PI(3)P. (A) GFP-FYVE domain from FENS-1 bound to PI(3)P-coupled lipid beads, whereas the GFP-ER–targeting domain of DFCP1 showed little binding (first four lanes; in, 5% of input; 3P, the faction bound to PI(3)P-coupled beads). A construct with the FENS-1 FYVE domain fused to the ER domain of DFCP1 (GFP-ERFYVE) maintained PI(3)P binding, whereas a construct with a point mutation in the FYVE domain (GFP-ERFYVE*) showed much-reduced binding. (B) Stable cell lines expressing the four constructs shown in A were left untreated or starved for 45 min before fixation and fluorescence microscopy. Note that the GFP-ERFYVE construct translocated to punctate structures upon starvation. (C) Cells expressing GFP-ERFYVE were starved alone or in the presence of wortmannin or BFA as indicated. (D) Cells expressing GFP-ERFYVE and starved for 45 min were either counterstained for the ER using antibodies to KDEL (top) or cotransfected with mRFP-MAP-LC3 (bottom). Note that the GFP-ERFYVE punctate structures frequently localized on the ER and showed partial colocalization with LC3. The panels on the right show an enlarged view of the boxed regions on the left. Bars, 10 μm.
Figure 4.
ER-anchored FYVE domain translocates to starvation-induced punctate structures in a pathway dependent on PI(3)P. (A) Three GFP-based constructs were fitted with a linker region, a transmembrane domain (TM), and a short cytoplasmic tail: GFP alone (GFP-TM), GFP fused to the WT FYVE domain of FENS-1 (GFP-TM-FYVE), and GFP fused to a point mutant of the FENS-1 FYVE domain unable to bind PI(3)P (GFP-FYVE*-TM). All three constructs were expressed transiently in HEK-293 cells, and their localization was examined with or without amino acid starvation for 60 min as indicated. Note that GFP-TM-FYVE translocated to punctate structures during starvation. (B) HEK-293 cells were transfected with GFP-FYVE-TM and mRFP-LC3 and starved for 60 min. Note that puncta of the two reporters colocalize, with the GFP construct frequently encircling mRFP-LC3 membranes (arrows). In general, cells had many more GFP-FYVE-TM than mRFP-LC3 puncta, and on average, 80% of LC3 puncta colocalized with a GFP-FYVE-TM particle. Insets show enlarged views of the boxed regions. (C) Live imaging of HEK-293 cells coexpressing GFP-FYVE-TM and dsRED-ER and starved for 60 min. Also see Video 1 (available at
http://www.jcb.org/cgi/content/full/jcb.200803137/DC1
). (D) A region indicated in C (bottom middle, box) is expanded and shown for 28–40 min during starvation. Note the formation and collapse of a ringlike particle. Arrows mark the particle at early stages. Bars: (A–C) 20 μm; (D) 1 μm.
Figure 5.
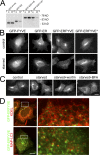
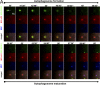
Isolation and characterization of stable HEK-293 clones expressing GFP-DFCP1. (A) Four clones were screened for GFP-DFCP1 expression (inset shows blots of endogenous DFCP1 and GFP-DFCP1 with β-COP as loading control; the fold overexpression of tagged DFCP1 over endogenous DFCP1 is also indicated) and for a good response to starvation, as measured by translocation of GFP-MAP-LC3 to punctate structures (shown in the graph) or acquisition of endogenous LC3-II form (last lane of inset). High levels of DFCP1 inhibited starvation responses (especially evident for clone 206). Error bars show the standard deviation from three independent experiments. (B) Clone 201 cells were left untreated or starved for 45 min in the absence or presence of wortmannin (wortm) or BFA as indicated, and the distribution of GFP-DFCP1 was examined by fluorescence microscopy. Note that wortmannin inhibited translocation, whereas BFA was without effect. (C) Clone 201 cells were treated with siRNA against Vps34 or beclin-1, or with a control siRNA as shown, starved for 60 min, and examined by fluorescence microscopy. Note that the level of punctate structures is reduced in Vps34- and beclin-1–reduced cells; this is analyzed for three independent experiments in D (error bars show standard deviation). (D, inset) The levels of Vps34 and beclin-1 after siRNA treatments. Of peripheral interest for this work is that the reduction of Vps34 during treatment with siRNA for beclin-1 is reproducible. (E) Clone 201 cells were starved for 60 min. Imaging was at 1 frame per 20 s, and selected frames throughout the sequence are shown. Also see Video 2 (available at
http://www.jcb.org/cgi/content/full/jcb.200803137/DC1
). (F) For a selected time interval (starting approximately at 35 min after starvation), a sequence showing formation and collapse of an omegasome (arrows) is shown. Bars (A–C and E) 20 μm; (F) 2 μm.
Figure 6.
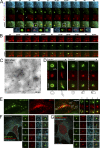
Relationship of PI(3)P-containing omegasomes with the ER and autophagosomes by live imaging and in fixed cells. (A) Clone 201 cells were transfected with mRFP-MAP-LC3 (red) and CFP-ER (blue) and starved for 60 min. Imaging was performed at 1 frame per 10 s, and a selected interval within this sequence is shown, starting at 33 min after starvation. Arrows indicate the first discernible omegasome (green) and autophagosome (red) occurrences. The enlarged panels are from the two boxed areas and represent views of (1) ER, (2) DFCP1, (3) MAP-LC3, (4) ER-MAPLC3, (5) DFCP1-MAPLC3, and (6) MAPLC3-ER, in that order. Also see Video 3 (available at
http://www.jcb.org/cgi/content/full/jcb.200803137/DC1
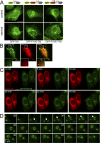
). (B) Dynamic relation of omegasomes and the ER using TIRFM imaging. Note that omegasomes form in regions containing ER and collapse there. Also see Video 4. (C) Thawed cryosection of clone 201 cells 45 min after starvation and double-labeled for anti–GFP-DFCPI (arrowheads, 10 nm gold) and anti-PDI (arrows, 5 nm gold). Note the putative autophagic-like vacuoles (asterisks) labeled for DFCPI-GFP adjacent to PDI-labeled ER membranes. This is one panel of a multipanel supplemental figure (Fig. S4). (D) Selected frames from live imaging of GFP-DFCP1 and mRFP-MAP-LC3 during starvation, whereby an intermediate is formed in which MAP-LC3 appears to bud from the omegasome while still being outlined with a DFCP1-staining membrane. At the bottom of each panel are line drawings of these structures using magnified photographs of the relevant frames. Also see Video 6. (E) Colocalization of omegasomes with a PI(3)P-binding protein applied exogenously. Clone 201 cells were starved for 60 min, perforated using nitrocellulose, and stained with purified GST-PX domain from p40phox. The majority of GSP-PX domain stains early endosomes (not depicted) but a substantial amount of the protein also binds to DFCP1 omegasomes (F and G). (F and G) Relationship of omegasomes to autophagy proteins and the ER. Clone 201 cells counterstained with antibodies to endogenous ER and endogenous MAP-LC3 or Atg5 as indicated. Selected examples from such cells (in addition to the one shown) where the ER was in a single layer and well resolved from cytosol, to allow evaluation of colocalizations, are shown in magnified panels 1–5. Bars: (A, B, and D) 1 μm; (E–G) 20 μm.
Figure 7.
Autophagosome maturation after omegasome exit. Cells expressing GFP-DFCP1 and mRFP-MAP-LC3 were starved and imaged for the indicated time interval. At 30 min after starvation, the cells were also incubated with 2 μM MDC. Note that an autophagosome emerges first from an omegasome (panels labeled autophagosome formation; arrow indicates the omegasome) and then begins to stain with MDC (panels labeled autophagosome maturation) without appearing to change its appearance or to fuse with another MDC-positive vesicle.
Figure 8.
Vps34 dynamics during omegasome formation. (A) Clone 201 cells were transfected with mRFP-Vps34, and a stable population expressing both proteins was selected (last three lanes). Note that exogenous mRFP-Vps34 was comparable to endogenous Vps34 and stable during amino acid starvation. (B) Cells as in A were starved for 60 min, fixed, and stained for Lamp-2. Note that DFCP1 punctate structures are frequently in proximity to Vps34 membranes (arrows on the bottom left) and that the majority but not all of Lamp-2 vesicles (∼80%) colocalize with Vps34, whereas all of the Vps34 vesicles colocalize with Lamp-2 (arrows on the bottom right indicate examples of Lamp-2 vesicles devoid of Vps34). (C) Selected frames from live imaging of mRFP-Vps34 in cells also expressing CFP-ER. Note that the Vps34 vesicle is in constant proximity to the ER and frequently appears to use the ER strands to move long distances (arrows on the bottom). The times refer to the period after amino acid starvation, but similar types of movement are evident without starvation. (D and E) Selected frames from live imaging of mRFP-Vps34, GFP-DFCP1, and CFP-ER during amino-acid starvation. (D) An omegasome being formed in constant and close proximity to a Vps34 particle. Bar, 1 μm. For selected frames of this video, as indicated (10–12 and 35–37), E shows the relationship of Vps34 to DFCP1 (top), Vps34 to ER (middle), and all three (bottom). Also see Video 7 (available at
http://www.jcb.org/cgi/content/full/jcb.200803137/DC1
). (F) An additional example from live imaging experiments showing an omegasome forming in close proximity to a Vps34 particle.
Figure 9.
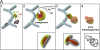
Potential role of omegasomes in autophagosome biogenesis. Our hypothesis is that during amino acid starvation, Vps34-containing vesicles interact with the ER and form PI(3)P on a membrane connected to the ER (step 1, green). This membrane domain associates with autophagosomal proteins (red) to create a mixed membrane domain that continues to expand but maintains its spatial separation with PI(3)P on the outside, and autophagosomal membranes inside (steps 1 and 2). Note that the connection of this membrane to the ER is not fully shown, thus the question marks at the junction. Once omegasomes reach their maximum size, the autophagosomal membranes bud inwards (steps 2 and 3), giving rise to a fully formed double-membrane autophagosome (step 4). The PI(3)P outer membrane in the intermediate of step 3 may aid the fusion reaction. Inward budding would allow the autophagosome to ingest cytoplasmic material and/or organelles indicated as autophagosome cargo. At the bottom of steps 2, 3, and 4, we have drawn cut-outs of the relevant structures to indicate the geometry of the bilayer.
Comment in
- Self-eating from an ER-associated cup.
Simonsen A, Stenmark H. Simonsen A, et al. J Cell Biol. 2008 Aug 25;182(4):621-2. doi: 10.1083/jcb.200807061. J Cell Biol. 2008. PMID: 18725534 Free PMC article.
Similar articles
- Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins.
Itakura E, Mizushima N. Itakura E, et al. Autophagy. 2010 Aug;6(6):764-76. doi: 10.4161/auto.6.6.12709. Autophagy. 2010. PMID: 20639694 Free PMC article. - WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1.
Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, Tooze SA. Dooley HC, et al. Mol Cell. 2014 Jul 17;55(2):238-52. doi: 10.1016/j.molcel.2014.05.021. Epub 2014 Jun 19. Mol Cell. 2014. PMID: 24954904 Free PMC article. - A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis.
Zhuang X, Wang H, Lam SK, Gao C, Wang X, Cai Y, Jiang L. Zhuang X, et al. Plant Cell. 2013 Nov;25(11):4596-615. doi: 10.1105/tpc.113.118307. Epub 2013 Nov 18. Plant Cell. 2013. PMID: 24249832 Free PMC article. - Biogenesis of autophagosomal precursors for LC3 lipidation from the ER-Golgi intermediate compartment.
Ge L, Wilz L, Schekman R. Ge L, et al. Autophagy. 2015;11(12):2372-4. doi: 10.1080/15548627.2015.1105422. Autophagy. 2015. PMID: 26565421 Free PMC article. Review. - WIPI2b and Atg16L1: setting the stage for autophagosome formation.
Wilson MI, Dooley HC, Tooze SA. Wilson MI, et al. Biochem Soc Trans. 2014 Oct;42(5):1327-34. doi: 10.1042/BST20140177. Biochem Soc Trans. 2014. PMID: 25233411 Review.
Cited by
- Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB.
Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, Montefusco S, Scotto-Rosato A, Prezioso C, Forrester A, Settembre C, Wang W, Gao Q, Xu H, Sandri M, Rizzuto R, De Matteis MA, Ballabio A. Medina DL, et al. Nat Cell Biol. 2015 Mar;17(3):288-99. doi: 10.1038/ncb3114. Nat Cell Biol. 2015. PMID: 25720963 Free PMC article. - This old heart: Cardiac aging and autophagy.
Linton PJ, Gurney M, Sengstock D, Mentzer RM Jr, Gottlieb RA. Linton PJ, et al. J Mol Cell Cardiol. 2015 Jun;83:44-54. doi: 10.1016/j.yjmcc.2014.12.017. Epub 2014 Dec 24. J Mol Cell Cardiol. 2015. PMID: 25543002 Free PMC article. Review. - Triggering mitophagy with far-red fluorescent photosensitizers.
Hsieh CW, Chu CH, Lee HM, Yuan Yang W. Hsieh CW, et al. Sci Rep. 2015 May 29;5:10376. doi: 10.1038/srep10376. Sci Rep. 2015. PMID: 26022357 Free PMC article. - Mitochondrial electron transport chain complex III is required for antimycin A to inhibit autophagy.
Ma X, Jin M, Cai Y, Xia H, Long K, Liu J, Yu Q, Yuan J. Ma X, et al. Chem Biol. 2011 Nov 23;18(11):1474-81. doi: 10.1016/j.chembiol.2011.08.009. Chem Biol. 2011. PMID: 22118681 Free PMC article. - Phosphatidylinositol-3-phosphate is light-regulated and essential for survival in retinal rods.
He F, Agosto MA, Anastassov IA, Tse DY, Wu SM, Wensel TG. He F, et al. Sci Rep. 2016 Jun 1;6:26978. doi: 10.1038/srep26978. Sci Rep. 2016. PMID: 27245220 Free PMC article.
References
- Asano, Y., H. Ihn, K. Yamane, M. Jinnin, Y. Mimura, and K. Tamaki. 2004. Phosphatidylinositol 3-kinase is involved in alpha2(I) collagen gene expression in normal and scleroderma fibroblasts. J. Immunol. 172:7123–7135. - PubMed
- Bampton, E.T., C.G. Goemans, D. Niranjan, N. Mizushima, and A.M. Tolkovsky. 2005. The dynamics of autophagy visualized in live cells: from autophagosome formation to fusion with endo/lysosomes. Autophagy. 1:23–36. - PubMed
- Blommaart, E.F., U. Krause, J.P. Schellens, H. Vreeling-Sindelarova, and A.J. Meijer. 1997. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur. J. Biochem. 243:240–246. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BBS/E/B/00001116/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/B/0000C116/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous