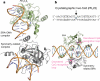The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix - PubMed (original) (raw)
The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix
Hideharu Hashimoto et al. Nature. 2008.
Abstract
Maintenance methylation of hemimethylated CpG dinucleotides at DNA replication forks is the key to faithful mitotic inheritance of genomic methylation patterns. UHRF1 (ubiquitin-like, containing PHD and RING finger domains 1) is required for maintenance methylation by interacting with DNA nucleotide methyltransferase 1 (DNMT1), the maintenance methyltransferase, and with hemimethylated CpG, the substrate for DNMT1 (refs 1 and 2). Here we present the crystal structure of the SET and RING-associated (SRA) domain of mouse UHRF1 in complex with DNA containing a hemimethylated CpG site. The DNA is contacted in both the major and minor grooves by two loops that penetrate into the middle of the DNA helix. The 5-methylcytosine has flipped completely out of the DNA helix and is positioned in a binding pocket with planar stacking contacts, Watson-Crick polar hydrogen bonds and van der Waals interactions specific for 5-methylcytosine. Hence, UHRF1 contains a previously unknown DNA-binding module and is the first example of a non-enzymatic, sequence-specific DNA-binding protein domain to use the base flipping mechanism to interact with DNA.
Figures
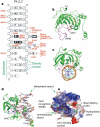
Figure 1. Structure of SRA–DNA complex
a, Summary of the SRA–DNA interactions; mc, main-chain-atom-mediated contacts; w, water-mediated hydrogen bonds. Black boxes represent CpG recognition sequence and K495-associated dotted lines represent weak hydrogen bonds. b, The side chains of V451 of the base flipping loop and R496 of the CpG recognition loop are in direct van der Waals contact. c, The two loops—CpG recognition and base flipping—penetrate into the DNA helix from opposite directions. d, The 5mC flips out and binds in a cage-like pocket. e, The surface charge at neutral pH is displayed as blue for positive (20 k_B_T), red for negative (−20 k_B_T), and white for neutral, where _k_B is the Boltzmann's constant and T is the temperature.
Figure 2. Details of SRA–DNA interactions
a, The 5mC•G base pair is shown in the front, and the adjoining G•C base pair is in the back. b, Planar stacking contacts of the extrahelical 5mC with Y471 and Y483 (left image). Omit electron densities, contoured at 4σ and 5σ above the mean, respectively, are shown for omitting 5mC (blue) or the methyl group (red) (right image). c, The hydrogen bond interactions with the polar atoms of 5mC. The double-dotted lines indicate van der Waals contacts with the methyl group of ring carbon C5. d, H450 forms a hydrogen bond from the minor groove side with cytosine of G•C pair at position 5 (see Fig. 1a). e, Network of internal polar interactions centred on residues H447 and S464. Gua, guanine. f, Network of internal charged interactions centred on residues R541 and D560. Distances are shown in angstroms.
Figure 3. Structure of the SRA–DNA specific (_P_212121) and non-specific (_P_6122) complexes
a, The crystal packing interactions in the _P_212121 space group may stabilize the DNA duplex. The extra contacts are made by the helix α4 of a symmetry-related molecule. These interactions are not present in the _P_41212 complex. b, A 12-base-pair duplex containing a single, centrally located unmethylated CG site was used to generate a non-specific complex with the SRA domain. A V451/H450-associated loop approaches the DNA from the minor groove at the junction of two head-to-head DNA molecules.
Similar articles
- Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism.
Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Arita K, et al. Nature. 2008 Oct 9;455(7214):818-21. doi: 10.1038/nature07249. Epub 2008 Sep 3. Nature. 2008. PMID: 18772891 - Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1.
Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S. Avvakumov GV, et al. Nature. 2008 Oct 9;455(7214):822-5. doi: 10.1038/nature07273. Epub 2008 Sep 3. Nature. 2008. PMID: 18772889 - The UHRF1 protein stimulates the activity and specificity of the maintenance DNA methyltransferase DNMT1 by an allosteric mechanism.
Bashtrykov P, Jankevicius G, Jurkowska RZ, Ragozin S, Jeltsch A. Bashtrykov P, et al. J Biol Chem. 2014 Feb 14;289(7):4106-15. doi: 10.1074/jbc.M113.528893. Epub 2013 Dec 24. J Biol Chem. 2014. PMID: 24368767 Free PMC article. - Coordinated Dialogue between UHRF1 and DNMT1 to Ensure Faithful Inheritance of Methylated DNA Patterns.
Bronner C, Alhosin M, Hamiche A, Mousli M. Bronner C, et al. Genes (Basel). 2019 Jan 18;10(1):65. doi: 10.3390/genes10010065. Genes (Basel). 2019. PMID: 30669400 Free PMC article. Review. - Targeting the SET and RING-associated (SRA) domain of ubiquitin-like, PHD and ring finger-containing 1 (UHRF1) for anti-cancer drug development.
Patnaik D, Estève PO, Pradhan S. Patnaik D, et al. Oncotarget. 2018 May 25;9(40):26243-26258. doi: 10.18632/oncotarget.25425. eCollection 2018 May 25. Oncotarget. 2018. PMID: 29899856 Free PMC article. Review.
Cited by
- Structural insight into the DNMT1 reaction cycle by cryo-electron microscopy.
De I, Weidenhausen J, Concha N, Müller CW. De I, et al. PLoS One. 2024 Sep 3;19(9):e0307850. doi: 10.1371/journal.pone.0307850. eCollection 2024. PLoS One. 2024. PMID: 39226277 Free PMC article. - The ICF syndrome protein CDCA7 harbors a unique DNA binding domain that recognizes a CpG dyad in the context of a non-B DNA.
Hardikar S, Ren R, Ying Z, Zhou J, Horton JR, Bramble MD, Liu B, Lu Y, Liu B, Coletta LD, Shen J, Dan J, Zhang X, Cheng X, Chen T. Hardikar S, et al. Sci Adv. 2024 Aug 23;10(34):eadr0036. doi: 10.1126/sciadv.adr0036. Epub 2024 Aug 23. Sci Adv. 2024. PMID: 39178265 Free PMC article. - CDCA7 is an evolutionarily conserved hemimethylated DNA sensor in eukaryotes.
Wassing IE, Nishiyama A, Shikimachi R, Jia Q, Kikuchi A, Hiruta M, Sugimura K, Hong X, Chiba Y, Peng J, Jenness C, Nakanishi M, Zhao L, Arita K, Funabiki H. Wassing IE, et al. Sci Adv. 2024 Aug 23;10(34):eadp5753. doi: 10.1126/sciadv.adp5753. Epub 2024 Aug 23. Sci Adv. 2024. PMID: 39178260 Free PMC article. - Structural analysis of the BisI family of modification dependent restriction endonucleases.
Szafran K, Rafalski D, Skowronek K, Wojciechowski M, Kazrani AA, Gilski M, Xu SY, Bochtler M. Szafran K, et al. Nucleic Acids Res. 2024 Aug 27;52(15):9103-9118. doi: 10.1093/nar/gkae634. Nucleic Acids Res. 2024. PMID: 39041409 Free PMC article. - Structure of human DPPA3 bound to the UHRF1 PHD finger reveals its functional and structural differences from mouse DPPA3.
Shiraishi N, Konuma T, Chiba Y, Hokazono S, Nakamura N, Islam MH, Nakanishi M, Nishiyama A, Arita K. Shiraishi N, et al. Commun Biol. 2024 Jun 19;7(1):746. doi: 10.1038/s42003-024-06434-9. Commun Biol. 2024. PMID: 38898124 Free PMC article.
References
- Bostick M, et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. - PubMed
- Sharif J, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. - PubMed
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. - PubMed
- Chen T, Li E. Establishment and maintenance of DNA methylation patterns in mammals. Curr. Top. Microbiol. Immunol. 2006;301:179–201. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 GM060398/GM/NIGMS NIH HHS/United States
- R01 GM049245-15/GM/NIGMS NIH HHS/United States
- R01 GM060398/GM/NIGMS NIH HHS/United States
- R01 GM049245/GM/NIGMS NIH HHS/United States
- GM060398/GM/NIGMS NIH HHS/United States
- GM049245/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- CA1263022/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases