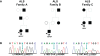Novel mutations in TARDBP (TDP-43) in patients with familial amyotrophic lateral sclerosis - PubMed (original) (raw)
. 2008 Sep 19;4(9):e1000193.
doi: 10.1371/journal.pgen.1000193.
Yong-Jie Zhang, Matt Baker, Jennifer M Gass, Nicole A Finch, Ya-Fei Xu, Heather Stewart, Brendan J Kelley, Karen Kuntz, Richard J P Crook, Jemeen Sreedharan, Caroline Vance, Eric Sorenson, Carol Lippa, Eileen H Bigio, Daniel H Geschwind, David S Knopman, Hiroshi Mitsumoto, Ronald C Petersen, Neil R Cashman, Mike Hutton, Christopher E Shaw, Kevin B Boylan, Bradley Boeve, Neill R Graff-Radford, Zbigniew K Wszolek, Richard J Caselli, Dennis W Dickson, Ian R Mackenzie, Leonard Petrucelli, Rosa Rademakers
Affiliations
- PMID: 18802454
- PMCID: PMC2527686
- DOI: 10.1371/journal.pgen.1000193
Novel mutations in TARDBP (TDP-43) in patients with familial amyotrophic lateral sclerosis
Nicola J Rutherford et al. PLoS Genet. 2008.
Abstract
The TAR DNA-binding protein 43 (TDP-43) has been identified as the major disease protein in amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration with ubiquitin inclusions (FTLD-U), defining a novel class of neurodegenerative conditions: the TDP-43 proteinopathies. The first pathogenic mutations in the gene encoding TDP-43 (TARDBP) were recently reported in familial and sporadic ALS patients, supporting a direct role for TDP-43 in neurodegeneration. In this study, we report the identification and functional analyses of two novel and one known mutation in TARDBP that we identified as a result of extensive mutation analyses in a cohort of 296 patients with variable neurodegenerative diseases associated with TDP-43 histopathology. Three different heterozygous missense mutations in exon 6 of TARDBP (p.M337V, p.N345K, and p.I383V) were identified in the analysis of 92 familial ALS patients (3.3%), while no mutations were detected in 24 patients with sporadic ALS or 180 patients with other TDP-43-positive neurodegenerative diseases. The presence of p.M337V, p.N345K, and p.I383V was excluded in 825 controls and 652 additional sporadic ALS patients. All three mutations affect highly conserved amino acid residues in the C-terminal part of TDP-43 known to be involved in protein-protein interactions. Biochemical analysis of TDP-43 in ALS patient cell lines revealed a substantial increase in caspase cleaved fragments, including the approximately 25 kDa fragment, compared to control cell lines. Our findings support TARDBP mutations as a cause of ALS. Based on the specific C-terminal location of the mutations and the accumulation of a smaller C-terminal fragment, we speculate that TARDBP mutations may cause a toxic gain of function through novel protein interactions or intracellular accumulation of TDP-43 fragments leading to apoptosis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Figure 1. Missense mutations identified in TARDBP in familial ALS patients.
(A) Pedigrees showing family history of ALS for three probands carrying TARDBP mutations. Black symbols represent patients affected with ALS; white symbols represent unaffected individuals. Pedigrees are constructed based on family history data provided by the NINDS Human Genetics Resource Center DNA and Cell Line Repository (
). The alive/dead status of individuals is unknown. Arrowheads indicate the probands. The onset age of ALS symptoms and the TARDBP mutation identified are included below each proband. (B) DNA sequence traces observed in a sample from the proband of each family. The observed single base substitution and predicted amino acid change are indicated below each chromatogram. cDNA numbering is according to the largest TARDBP transcript (NM_007375.3) and starting at the translation initiation codon. Protein numbering is relative to the largest TDP-43 isoform (NP_031401.1).
Figure 2. Overview of mutations identified to date in TARDBP.
Schematic overview of the 7 TARDBP exons showing coding regions in dark blue and non-coding regions in light blue (top). The TDP-43 protein structure with location of the conserved domains is shown with protein numbering according to the largest isoforms NP_031401.1 (middle). Protein sequence alignment shows strong conservation in the C-terminal region of TDP-43 (bottom). Colored boxes indicate the position of known and novel TDP-43 mutations identified in sporadic (orange) and familial (red) ALS patients. TDP-43 mutations identified in this study are underlined. Orange and red lines in TARDBP gene and TDP-43 protein indicate approximate positions of the mutations. RRM = RNA recognition motif.
Figure 3. Biochemical analysis of TDP-43 in lymphoblastoid cell lines of TARDBP mutation carriers.
Western blot analyses of protein lysates derived from lymphoblastoid cell lines from 3 familial ALS patients carrying different TARDBP mutations (p.M337V, p.N345K and p.I383V), 2 ALS patients (1 and 2) without TARDBP mutations and 5 healthy control individuals (Control 1–5). In lymphoblastoid cell lines derived from TARDBP mutation carriers and sporadic ALS patients an accumulation of 2 smaller C-terminal fragments of TDP-43 protein of approximately 35 and 25 kDa was observed in detergent-insoluble fractions treated with the proteasome inhibitor, MG-132. In lymphoblastoid cell lines derived from control individuals the levels of the 35 kDa fragment were substantially lower, and the 25 kDa fragment was mostly undetectable. Membranes from the soluble fraction were reprobed for beta-actin to monitor protein loading.
Figure 4. Proteasome inhibition increases the proteolytic cleavage of TDP-43.
Western blot analyses of H4 neuroglioma cells treated with the proteasome inhibitor, PSI (10 µM, 24 hours) and a pan-caspase inhibitor, Z-VAD-FMK (100 µM, 24 hours) separately or in combination. Treatment with PSI revealed an increase in proteolytic cleavage of TDP-43 fragments (35 and 25 kDa) and an increase in caspase-3 activity. Treatment with a pan-caspase inhibitor suppressed PSI-induced TDP-43 cleavage and caspase-3 activity. HSP70 levels were increased after PSI treatment and the levels persisted in the presence of a pan-caspase inhibitor. Similar results were obtained in 3 independent experiments.
Similar articles
- TARDBP mutation analysis in TDP-43 proteinopathies and deciphering the toxicity of mutant TDP-43.
Gendron TF, Rademakers R, Petrucelli L. Gendron TF, et al. J Alzheimers Dis. 2013;33 Suppl 1(Suppl 1):S35-45. doi: 10.3233/JAD-2012-129036. J Alzheimers Dis. 2013. PMID: 22751173 Free PMC article. Review. - TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis.
Van Deerlin VM, Leverenz JB, Bekris LM, Bird TD, Yuan W, Elman LB, Clay D, Wood EM, Chen-Plotkin AS, Martinez-Lage M, Steinbart E, McCluskey L, Grossman M, Neumann M, Wu IL, Yang WS, Kalb R, Galasko DR, Montine TJ, Trojanowski JQ, Lee VM, Schellenberg GD, Yu CE. Van Deerlin VM, et al. Lancet Neurol. 2008 May;7(5):409-16. doi: 10.1016/S1474-4422(08)70071-1. Epub 2008 Apr 7. Lancet Neurol. 2008. PMID: 18396105 Free PMC article. - TARDBP (TDP-43) sequence analysis in patients with familial and sporadic ALS: identification of two novel mutations.
Del Bo R, Ghezzi S, Corti S, Pandolfo M, Ranieri M, Santoro D, Ghione I, Prelle A, Orsetti V, Mancuso M, Sorarù G, Briani C, Angelini C, Siciliano G, Bresolin N, Comi GP. Del Bo R, et al. Eur J Neurol. 2009 Jun;16(6):727-32. doi: 10.1111/j.1468-1331.2009.02574.x. Epub 2009 Feb 19. Eur J Neurol. 2009. PMID: 19236453 - A novel TARDBP insertion/deletion mutation in the flail arm variant of amyotrophic lateral sclerosis.
Solski JA, Yang S, Nicholson GA, Luquin N, Williams KL, Fernando R, Pamphlett R, Blair IP. Solski JA, et al. Amyotroph Lateral Scler. 2012 Sep;13(5):465-70. doi: 10.3109/17482968.2012.662690. Epub 2012 Mar 16. Amyotroph Lateral Scler. 2012. PMID: 22424122 - ALS and FTLD: two faces of TDP-43 proteinopathy.
Liscic RM, Grinberg LT, Zidar J, Gitcho MA, Cairns NJ. Liscic RM, et al. Eur J Neurol. 2008 Aug;15(8):772-80. doi: 10.1111/j.1468-1331.2008.02195.x. Eur J Neurol. 2008. PMID: 18684309 Free PMC article. Review.
Cited by
- U1 snRNP is mislocalized in ALS patient fibroblasts bearing NLS mutations in FUS and is required for motor neuron outgrowth in zebrafish.
Yu Y, Chi B, Xia W, Gangopadhyay J, Yamazaki T, Winkelbauer-Hurt ME, Yin S, Eliasse Y, Adams E, Shaw CE, Reed R. Yu Y, et al. Nucleic Acids Res. 2015 Mar 31;43(6):3208-18. doi: 10.1093/nar/gkv157. Epub 2015 Mar 3. Nucleic Acids Res. 2015. PMID: 25735748 Free PMC article. - The phosphatase calcineurin regulates pathological TDP-43 phosphorylation.
Liachko NF, Saxton AD, McMillan PJ, Strovas TJ, Currey HN, Taylor LM, Wheeler JM, Oblak AL, Ghetti B, Montine TJ, Keene CD, Raskind MA, Bird TD, Kraemer BC. Liachko NF, et al. Acta Neuropathol. 2016 Oct;132(4):545-61. doi: 10.1007/s00401-016-1600-y. Epub 2016 Jul 29. Acta Neuropathol. 2016. PMID: 27473149 Free PMC article. - Interleukin-6 Deficiency Does Not Affect Motor Neuron Disease Caused by Superoxide Dismutase 1 Mutation.
Han Y, Ripley B, Serada S, Naka T, Fujimoto M. Han Y, et al. PLoS One. 2016 Apr 12;11(4):e0153399. doi: 10.1371/journal.pone.0153399. eCollection 2016. PLoS One. 2016. PMID: 27070121 Free PMC article. - Wild type TDP-43 induces neuro-inflammation and alters APP metabolism in lentiviral gene transfer models.
Herman AM, Khandelwal PJ, Rebeck GW, Moussa CE. Herman AM, et al. Exp Neurol. 2012 May;235(1):297-305. doi: 10.1016/j.expneurol.2012.02.011. Epub 2012 Feb 28. Exp Neurol. 2012. PMID: 22402344 Free PMC article. - RNA processing defects associated with diseases of the motor neuron.
Kolb SJ, Sutton S, Schoenberg DR. Kolb SJ, et al. Muscle Nerve. 2010 Jan;41(1):5-17. doi: 10.1002/mus.21428. Muscle Nerve. 2010. PMID: 19697368 Free PMC article. Review.
References
- Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. - PubMed
- Davidson Y, Kelley T, Mackenzie IR, Pickering-Brown S, Du Plessis D, et al. Ubiquitinated pathological lesions in frontotemporal lobar degeneration contain the TAR DNA-binding protein, TDP-43. Acta Neuropathol. 2007;113:521–533. - PubMed
- Kwong LK, Uryu K, Trojanowski JQ, Lee VM. TDP-43 proteinopathies: neurodegenerative protein misfolding diseases without amyloidosis. Neurosignals. 2008;16:41–51. - PubMed
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS42759-04S1/NS/NINDS NIH HHS/United States
- P50 NS040256/NS/NINDS NIH HHS/United States
- P30 AG13854/AG/NIA NIH HHS/United States
- P50 AG016574/AG/NIA NIH HHS/United States
- P50 NS 40256/NS/NINDS NIH HHS/United States
- P30 AG013854-139003/AG/NIA NIH HHS/United States
- P01 AG17216/AG/NIA NIH HHS/United States
- R01 AG015866/AG/NIA NIH HHS/United States
- P01 NS040256/NS/NINDS NIH HHS/United States
- P01 AG03949/AG/NIA NIH HHS/United States
- P50 AG25711/AG/NIA NIH HHS/United States
- P01 AG003949/AG/NIA NIH HHS/United States
- U01 AG006786/AG/NIA NIH HHS/United States
- R01 NS042759/NS/NINDS NIH HHS/United States
- P30 AG013854/AG/NIA NIH HHS/United States
- P50 AG16574/AG/NIA NIH HHS/United States
- R01 NS42759/NS/NINDS NIH HHS/United States
- U01 AG06786/AG/NIA NIH HHS/United States
- NS40256/NS/NINDS NIH HHS/United States
- G0500289/MRC_/Medical Research Council/United Kingdom
- AG19610-01/AG/NIA NIH HHS/United States
- P50 AG025711/AG/NIA NIH HHS/United States
- P30 AG019610/AG/NIA NIH HHS/United States
- R01 AG026251/AG/NIA NIH HHS/United States
- P01 AG017216/AG/NIA NIH HHS/United States
- R01 AG026251-01/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous