ROS3 is an RNA-binding protein required for DNA demethylation in Arabidopsis - PubMed (original) (raw)
. 2008 Oct 30;455(7217):1259-62.
doi: 10.1038/nature07305. Epub 2008 Sep 24.
Affiliations
- PMID: 18815596
- PMCID: PMC2782394
- DOI: 10.1038/nature07305
ROS3 is an RNA-binding protein required for DNA demethylation in Arabidopsis
Xianwu Zheng et al. Nature. 2008.
Abstract
DNA methylation is an important epigenetic mark for transcriptional gene silencing (TGS) in diverse organisms. Recent studies suggest that the methylation status of a number of genes is dynamically regulated by methylation and demethylation. In Arabidopsis, active DNA demethylation is mediated by the ROS1 (repressor of silencing 1) subfamily of 5-methylcytosine DNA glycosylases through a base excision repair pathway. These demethylases have critical roles in erasing DNA methylation and preventing TGS of target genes. However, it is not known how the demethylases are targeted to specific sequences. Here we report the identification of ROS3, an essential regulator of DNA demethylation that contains an RNA recognition motif. Analysis of ros3 mutants and ros1 ros3 double mutants suggests that ROS3 acts in the same genetic pathway as ROS1 to prevent DNA hypermethylation and TGS. Gel mobility shift assays and analysis of ROS3 immunoprecipitate from plant extracts shows that ROS3 binds to small RNAs in vitro and in vivo. Immunostaining shows that ROS3 and ROS1 proteins co-localize in discrete foci dispersed throughout the nucleus. These results demonstrate a critical role for ROS3 in preventing DNA hypermethylation and suggest that DNA demethylation by ROS1 may be guided by RNAs bound to ROS3.
Figures
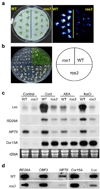
Figure 1. The ros3 mutation causes transcriptional gene silencing
a, Stress-induced expression of the RD29A-LUC transgene in wild type (WT) and ros3 mutant plants after treatment with 300 mM NaCl for 5 hr. b, Like ros1, the ros3 mutant plants are sensitive to kanamycin. c, Northern blots showing that ros3 reduces the transcript levels of LUC, NPTII and endogenous RD29A, but not of the control, COR15A. Plants were either untreated (Control) or treated with cold (4°C) for 24 hr, 100 µM ABA for 3 hr, or 300 mM NaCl for 5 hr. d, Nuclear run-on assay showing the pre-mRNA levels of LUC, NPTII and RD29A genes in wild-type and ros3. COR15A and CBF3 were used as controls.
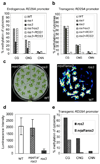
Figure 2. DNA hypermethylation in ros3 and suppression of ros3 by nrpd1a
a and b Bisulfite sequencing analysis of promoter methylation status of the endogenous RD29A (a) and RD29A-LUC transgene (b) in WT, ros1, ros3, ros1ros3, ros1 complemented with wild type ROS1 transgene, and ros3 complemented with wild type ROS3 transgene. c, Suppression of ros3 by the nrpd1a-1 mutation. Seedlings of WT, ros3 and nrpd1aros3 double mutant grown in MS medium for three weeks were transferred to a filter paper soaked with 300 mM NaCl for 5 hr. Left, picture of the seedlings; right, luminescence image. d, Quantification of luminescence in (c). Error bars represent standard deviation (n=20). e, DNA methylation status at the transgene RD29A promoter in ros3 and nrpd1aros3 plants.
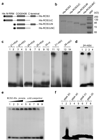
Figure 3. ROS3 binds small RNAs
a, Diagram of ROS3 and its truncated mutant forms. b, Coomassie stained SDS-PAGE gel showing the recombinant proteins used for RNA binding assays. c, ROS3 but not ROS3ΔNC binds 21-,24- and 26-nt single stranded RNA of sequence b. Lanes 1, 3, 5, 7, 9, 11 and 13, ROS3; lanes 2, 4, 6, 8, 10, 12 and 14, ROS3ΔNC. d, ROS3ΔC but not ROS3ΔN or ROS3ΔNC binds 24-nt single stranded RNA of sequence b. Lane 1, ROS3; lane 2, ROS3ΔN; lane 3, ROS3ΔC; lane 4, ROS3ΔNC. e, Protein concentration-dependent binding to 24-nt single stranded RNA (b) and competition by unlabeled small RNA. Lanes 1–4, increasing binding to 24-nt (b) small RNA by increasing ROS3 protein concentration; lanes 5–9, competition by increasing amount of cold 24-nt RNA (b). f, ROS3 binds 25-nt RNA (c) and RNA (d) in vitro. RNAs of sequences b and a are used as controls. Lanes 1, 3, 5 and 7, ROS3; lanes 2, 4, 6 and 8, ROS3ΔN.
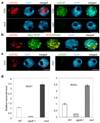
Figure 4. Co-localization of ROS3 with ROS1 in the nucleus of Arabidopsis mesophyll cells and assay of ROS1 and ROS3 mRNA levels
a, Localization of ROS3 and ROS1 by their respective antibodies. b, Dual immunolocalization of ROS3 and ROS1 with use of anti-ROS3 and anti-Myc. c, ROS1 localization in ros3 mutant and ROS3 localization in ros1 showing their inter-dependence for appropriate localization. Size bars correspond to 5 µm. d, Quantitative RT-PCR assay of relative ROS1 and ROS3 transcript levels in the various genotypes.
Similar articles
- Involvement of MEM1 in DNA demethylation in Arabidopsis.
Lu Y, Dai J, Yang L, La Y, Zhou S, Qiang S, Wang Q, Tan F, Wu Y, Kong W, La H. Lu Y, et al. Plant Mol Biol. 2020 Feb;102(3):307-322. doi: 10.1007/s11103-019-00949-0. Epub 2020 Jan 4. Plant Mol Biol. 2020. PMID: 31902068 - Regulatory link between DNA methylation and active demethylation in Arabidopsis.
Lei M, Zhang H, Julian R, Tang K, Xie S, Zhu JK. Lei M, et al. Proc Natl Acad Sci U S A. 2015 Mar 17;112(11):3553-7. doi: 10.1073/pnas.1502279112. Epub 2015 Mar 2. Proc Natl Acad Sci U S A. 2015. PMID: 25733903 Free PMC article. - Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation.
Agius F, Kapoor A, Zhu JK. Agius F, et al. Proc Natl Acad Sci U S A. 2006 Aug 1;103(31):11796-801. doi: 10.1073/pnas.0603563103. Epub 2006 Jul 24. Proc Natl Acad Sci U S A. 2006. PMID: 16864782 Free PMC article. - RNA-directed DNA methylation and Pol IVb in Arabidopsis.
Matzke M, Kanno T, Huettel B, Daxinger L, Matzke AJ. Matzke M, et al. Cold Spring Harb Symp Quant Biol. 2006;71:449-59. doi: 10.1101/sqb.2006.71.028. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381327 Review. - Preventing transcriptional gene silencing by active DNA demethylation.
Kapoor A, Agius F, Zhu JK. Kapoor A, et al. FEBS Lett. 2005 Oct 31;579(26):5889-98. doi: 10.1016/j.febslet.2005.08.039. Epub 2005 Aug 31. FEBS Lett. 2005. PMID: 16162337 Review.
Cited by
- Base Excision DNA Repair in Plants: Arabidopsis and Beyond.
Grin IR, Petrova DV, Endutkin AV, Ma C, Yu B, Li H, Zharkov DO. Grin IR, et al. Int J Mol Sci. 2023 Sep 29;24(19):14746. doi: 10.3390/ijms241914746. Int J Mol Sci. 2023. PMID: 37834194 Free PMC article. Review. - Recent Advances on DNA Base Flipping: A General Mechanism for Writing, Reading, and Erasing DNA Modifications.
Ren R, Horton JR, Hong S, Cheng X. Ren R, et al. Adv Exp Med Biol. 2022;1389:295-315. doi: 10.1007/978-3-031-11454-0_12. Adv Exp Med Biol. 2022. PMID: 36350515 Free PMC article. - Fine mapping and characterization of RLL6 locus required for anti-silencing of a transgene and DNA demethylation in Arabidopsis thaliana.
Wang X, Wang M, Dai J, Wang Q, La H. Wang X, et al. Front Genet. 2022 Sep 26;13:1008700. doi: 10.3389/fgene.2022.1008700. eCollection 2022. Front Genet. 2022. PMID: 36226182 Free PMC article. - Conditional GWAS of non-CG transposon methylation in Arabidopsis thaliana reveals major polymorphisms in five genes.
Sasaki E, Gunis J, Reichardt-Gomez I, Nizhynska V, Nordborg M. Sasaki E, et al. PLoS Genet. 2022 Sep 9;18(9):e1010345. doi: 10.1371/journal.pgen.1010345. eCollection 2022 Sep. PLoS Genet. 2022. PMID: 36084135 Free PMC article. - Gadd45 in DNA Demethylation and DNA Repair.
Chandramouly G. Chandramouly G. Adv Exp Med Biol. 2022;1360:55-67. doi: 10.1007/978-3-030-94804-7_4. Adv Exp Med Biol. 2022. PMID: 35505162
References
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. - PubMed
- Martienssen RA, Colot V. DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science. 2001;293:1070–1074. - PubMed
- Tariq M, Paszkowski J. DNA and histone methylation in plants. Trends Genet. 2004;20:244–251. - PubMed
- Bender J. DNA methylation and epigenetics. Annu. Rev. Plant Biol. 2004;55:41–68. - PubMed
- Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 2005;6:24–35. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01GM059138/GM/NIGMS NIH HHS/United States
- R01 GM070795/GM/NIGMS NIH HHS/United States
- R01GM077590/GM/NIGMS NIH HHS/United States
- R01 GM060380-08/GM/NIGMS NIH HHS/United States
- R01 GM077590-03/GM/NIGMS NIH HHS/United States
- R01 GM059138/GM/NIGMS NIH HHS/United States
- R01 GM060380/GM/NIGMS NIH HHS/United States
- R01GM070795/GM/NIGMS NIH HHS/United States
- R01 GM077590/GM/NIGMS NIH HHS/United States
- 1R01GM060380/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases