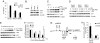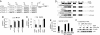AP4 encodes a c-MYC-inducible repressor of p21 - PubMed (original) (raw)
AP4 encodes a c-MYC-inducible repressor of p21
Peter Jung et al. Proc Natl Acad Sci U S A. 2008.
Abstract
In the majority of human tumors, expression of the c-MYC oncogene becomes constitutive. Here, we report that c-MYC directly regulates the expression of AP4 via CACGTG motifs in the first intron of the AP4 gene. Induction of AP4 was required for c-MYC-mediated cell cycle reentry of anti-estrogen arrested breast cancer cells and mitogen-mediated repression of the CDK inhibitor p21. AP4 directly repressed p21 by occupying four CAGCTG motifs in the p21 promoter via its basic region. AP4 levels declined after DNA damage, and ectopic AP4 interfered with p53-mediated cell cycle arrest and sensitized cells to apoptosis induced by DNA damaging agents. AP4 expression blocked induction of p21 by TGF-beta in human keratinocytes and interfered with up-regulation of p21 and cell cycle arrest during monoblast differentiation. Notably, AP4 is specifically expressed in colonic progenitor and colorectal carcinoma cells. In conclusion, our results indicate that c-MYC employs AP4 to maintain cells in a proliferative, progenitor-like state.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
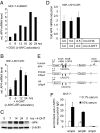
Characterization of AP4 as a direct c-MYC target gene. (A) Quantification of AP4 mRNA after activation of c-MYC. MCF-7-PJMMR1 cells were treated with ICI (1 μM) for 60 h before activation of c-MYC by addition of doxycycline (DOX, 1 μg/ml) for the indicated periods, and RNA was subjected to qPCR analysis. (B) Quantification of AP4 mRNA after c-MYC activation. HDF-MYC-ER cells were serum-deprived for 48 h. After addition of 4-OHT (200 nM), total RNA was isolated at the indicated time points from biological triplicates. AP4 mRNA expression was determined by qPCR analysis. Error bars indicate standard deviations. (C) AP4 protein expression after c-MYC activation. Protein lysates were prepared from HDF-MYC-ER cells at the indicated time points. Expression of AP4 and β-actin was determined by immunoblotting. (D) HDF-MYC-ER cells were grown to confluence and treated with 4-OHT (200 nM) and CHX (70 nM), as indicated. The expression level of AP4 after combined CHX/4-OHT treatment was normalized to cells treated with CHX alone. Expression of AP4 and, for normalization, β-actin mRNA was determined by qPCR. Analyses were performed in triplicates. Error bars indicate standard deviations. (E) Comparison of the mouse, rat, and human AP4 promoter regions. “+1” indicates the transcription start site. “amp” indicates PCR amplicons used for qChIP analysis with their positions relative to the transcription start site. Arrows indicate the approximate positions of canonical c-MYC-binding sites (CACGTG). The positions of these sites relative to the transcription start site (“+1”) are +660, +1262, +1645, and +1766 for human AP4; +560 and +1620 for the mouse tcfap4; and +666 and +1725 for the rat tcfap4, respectively. (F) Detection of c-MYC at the AP4 promoter. MCF-7 cells were serum-starved (0.1% serum) for 48 h or restimulated (10% serum) for 12 h. Chromatin was cross-linked and subjected to qChIP analysis with a c-MYC-specific antibody and, as a control, rabbit IgG. qPCR analysis was performed with primers flanking three of the four canonical E-boxes in the first AP4 intron (“ampA” and “ampB”; see also Fig. 1_C_) or a control primer pair (“ampC”) localized in the last intron of AP4. For normalization, a fragment not containing E-boxes from chromosome 16q22 was used.
Fig. 2.
Effects of AP4 on cell cycle progression and p21 expression. (A) Flow cytometric analysis of ICI (1 μM)-treated MCF-7-PJMMR1 cells after siRNA-mediated down-regulation of AP4. c-MYC was activated by addition of DOX (1 μg/ml) for 22 h. The depicted diagram shows the percentage of cells in S-phase after treatment with ICI alone or in combination with c-MYC overexpression (ICI+MYC). The experiment was performed in duplicate. The standard error is depicted. (B) MCF-7 cells were transfected with two different siRNAs targeting AP4 or a nonsilencing control siRNA. Expression of AP4, p21 and β-actin was detected by immunoblot analysis. (C) MCF-7 cells were serum-deprived for 48 h, restimulated with 10% serum for the indicated periods, and analyzed by immunoblotting. (D) MCF-7 cells were transfected with siRNAs targeting AP4 or a nonsilencing siRNA. Thirty-six hours later, cells were serum-starved for 30 h. After restimulation with 10% FBS-containing medium for the indicated periods, cell lysates were subjected to immunoblot analysis. (E) Expression of AP4-VSV, p53, p21, and β-actin proteins was detected by immunoblot analysis after induction of a conditional VSV-tagged AP4 allele in U-2OS cells. (F) Quantification of p21 mRNA after activation of AP4 in U-2OS cells by qPCR analysis. Expression of p21 was normalized to β-actin expression. (G) The proximal promoter region of the human p21 gene contains four AP4 binding sites (CAGCTG). “+1” indicates the transcription start site. “amp:” PCR amplicons used for qChIP analysis with their position relative to the transcription start site. Positions of two p53-binding sites (p53BDS) are indicated. The approximate positions of four putative AP4-binding sites (arrows) and the initiator (Inr) element (TCAGTTCCT) (filled square) are indicated (their precise positions relative to the transcription start site of p21 is depicted in Fig. 3_A_). (H) qChIP analysis of AP4 at the p21 promoter. A conditional VSV-tagged AP4 allele was induced by addition of DOX (100 ng/ml) for 16 h in U-2OS cells. Genomic DNA coprecipitated with an anti-VSV or mouse IgG antibody was analyzed by qPCR. For normalization, a fragment on chromosome 16q22, not containing E-boxes was used.
Fig. 3.
AP4 directly represses p21 expression via E-box motifs. (A) Schematic presentation of putative AP4-binding sites (A1-A4) and their position in the p21 promoter region relative to the transcriptional start site (+1). Wild-type and mutant p21 promoter constructs used in transient reporter assays are depicted. Mutated AP4-binding sites are represented in bold and underlined. The initiator (Inr) element (TCAGTTCCT) localizes to position + 8 to + 16 relative to the transcription start site (“+1”) (13). luc: ORF encoding the firefly luciferase. (B) Determination of p21 reporter activity in H1299 cells. Cells were transfected in 12-well plates with wild type or the indicated mutant p21 reporter plasmids, pcDNA3-AP4-VSV plasmid or equimolar amounts of pcDNA3-VSV backbone. Shown are the median expression values and standard errors of two independent transfection experiments. p21 prom. wt, mA3 + 4, mA2–4, and mA1–4: reporter plasmids encoding for the p21 promoter sequence with wild-type or mutant AP4-binding sites (see Fig. 3_A_). (C) The CMV/p21 reporter (nucleotides −49 to +16 of p21), a mutant version (CMV/p21mut) harboring substitutions of two nucleotides within three potential Miz1-binding sequences, or the p21/CMV reporter (nucleotides −94/-50 of p21) containing one E2F and four Sp1/3-binding sites (13) were cotransfected with pcDNA3-AP4-VSV plasmid or equimolar amounts of pcDNA3-VSV backbone in H1299 cells. Shown are the median expression values and standard errors of two independent transfection experiments. (D) Schematic representation of AP4 mutants. B: Basic region, HLH: helix–loop–helix, LZ1/2: leuzine zipper motif 1 and 2, TIV: conserved motif of unknown function containing the amino acid sequence TIV. The amino acid sequence of the basic region (underlined) and flanking residues are indicated for the wild type and mutant AP4 versions. Altered residues are represented in bold. (E) Effect of AP4 variants on p21 reporter activity in H1299 cells. Cells were transfected in 12-well plates with wild-type p21 reporter plasmid, pcDNA3-AP4-VSV plasmids (wild type or mutant AP4 versions) or equimolar amounts of pcDNA3-VSV backbone. Shown are the median expression values and standard deviations of three independent experiments. (F) Effect of AP4 variants on endogenous p21 expression. Expression level of AP4-VSV, p21, and β-actin proteins was detected by immunoblot analysis 24 h after induction of conditional wild-type or mutant AP4 alleles in U-2OS cells.
Fig. 4.
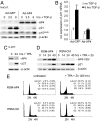
Role of AP4 in the DNA damage response. (A) Effect of DNA damage on AP4 expression. MCF-7 cells were treated with etoposide (ETOP, 20 μg/ml) and cell extracts were obtained at the indicated time points. Expression of the indicated proteins was determined by immunoblotting. (B) Ectopic AP4-VSV was induced in U-2OS cells for 12 h by addition of DOX (100 ng/ml). Then ETOP (20 μg/ml) was added for the indicated periods. Expression of the differentially phosphorylated retinoblastoma protein (Rb-P/Rb), AP4-VSV, p21 or β-actin was detected by immunoblotting. (C) p21 reporter activity was determined in H1299 cells transfected with the indicated plasmids. Increasing p53 expression was achieved by transfection of 0, 50 or 200 ng of plasmids (indicated as  ). Shown are the median expression values and standard errors of two independent transfection experiments. p21 mA3 + 4: see Fig. 3_A_. (D) AP4 was induced by DOX for 12 h before treatment of cells with ETOP (20 μg/ml) for 48 h. Then cells were analyzed by flow cytometry. Depicted are exemplary histograms representing 10,000 cells. 2N: cells in G1, 4N: cells in G2/M.
). Shown are the median expression values and standard errors of two independent transfection experiments. p21 mA3 + 4: see Fig. 3_A_. (D) AP4 was induced by DOX for 12 h before treatment of cells with ETOP (20 μg/ml) for 48 h. Then cells were analyzed by flow cytometry. Depicted are exemplary histograms representing 10,000 cells. 2N: cells in G1, 4N: cells in G2/M.
Fig. 5.
AP4 antagonizes TGF-β and TPA mediated p21 induction. (A) HaCaT cells were infected with adenovirus encoding AP4 and eGFP (Ad-AP4) or eGFP alone (Ad-GFP). Twenty-four hours later, cells were treated with human, recombinant TGF-β (5 ng/ml) for the indicated periods. Expression of AP4-VSV, p21, p15Ink4b and β-actin was determined by immunoblotting. (B) Quantification of p21 mRNA in HaCaT cells infected with adenoviruses encoding either AP4 and eGFP or eGFP alone. Twenty-four hours after infection, cells were treated with human recombinant TGF-β (5 ng/ml) for 6 h. mRNA expression of p21 and β-actin was determined by qPCR analysis. The experiment was performed in duplicate. Error bars indicate standard errors. (C) U-937 cells were treated with 10 nM TPA (10 nM) for 24 h, and expression of AP4, p21, and, as a control for equal loading, β-actin was determined by immunoblotting. (D) U-937 RSM-AP4 or RSM-Ctrl cell pools were treated with 10 nM TPA and 100 μM zinc sulfate for the indicated periods. Expression of AP4-VSV, p21 and β-actin was determined by immunoblotting. (E) U-937 RSM-AP4 or RSM-Ctrl cells were treated with 10 nM TPA and 100 μM zinc sulfate for the indicated periods, and analyzed by flow cytometry. The experiment was repeated twice, and exemplary histograms representing 15,000 cells are shown. The percentage cell cycle distribution represent the average of two independent experiments. Standard errors are indicated. 2N: cells in G1, 4N: cells in G2/M.
Fig. 6.
AP4 expression in human colon and colorectal cancer. (A) Section of an endoscopic biopsy derived from a normal colonic region. Consecutive paraffin sections were stained with antibodies directed against Ki67, AP4 or p21 (arrows indicate the presence of positive cells). Magnification: ×200. (B) Sections of endoscopic biopsies derived from primary tumors of two patients are depicted. Consecutive paraffin sections were stained with antibodies directed against c-MYC, Ki67, AP4, or p21. Identical results were obtained with colorectal carcinoma biopsies from 10 additional patients (see
Fig. S9
). A, adenomatous polyp (dysplastic); T, tumor. Magnification: ×200.
Similar articles
- Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage.
Seoane J, Le HV, Massagué J. Seoane J, et al. Nature. 2002 Oct 17;419(6908):729-34. doi: 10.1038/nature01119. Epub 2002 Oct 2. Nature. 2002. PMID: 12384701 - Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter.
Wu S, Cetinkaya C, Munoz-Alonso MJ, von der Lehr N, Bahram F, Beuger V, Eilers M, Leon J, Larsson LG. Wu S, et al. Oncogene. 2003 Jan 23;22(3):351-60. doi: 10.1038/sj.onc.1206145. Oncogene. 2003. PMID: 12545156 - Role of c-myc regulation in Zta-mediated induction of the cyclin-dependent kinase inhibitors p21 and p27 and cell growth arrest.
Rodriguez A, Jung EJ, Yin Q, Cayrol C, Flemington EK. Rodriguez A, et al. Virology. 2001 Jun 5;284(2):159-69. doi: 10.1006/viro.2001.0923. Virology. 2001. PMID: 11384216 - Functions of myc:max in the control of cell proliferation and tumorigenesis.
Hurlin PJ, Dezfouli S. Hurlin PJ, et al. Int Rev Cytol. 2004;238:183-226. doi: 10.1016/S0074-7696(04)38004-6. Int Rev Cytol. 2004. PMID: 15364199 Review. - Transcriptional regulation and transformation by Myc proteins.
Adhikary S, Eilers M. Adhikary S, et al. Nat Rev Mol Cell Biol. 2005 Aug;6(8):635-45. doi: 10.1038/nrm1703. Nat Rev Mol Cell Biol. 2005. PMID: 16064138 Review.
Cited by
- AP4 induces JNK1 and a miR-22-3p/FOSL1 feed-forward loop to activate AP-1 and promote colorectal cancer metastasis.
Chou J, Kaller M, Rokavec M, Liu F, Hermeking H. Chou J, et al. Cancer Commun (Lond). 2024 Mar;44(3):433-437. doi: 10.1002/cac2.12514. Epub 2024 Jan 15. Cancer Commun (Lond). 2024. PMID: 38225895 Free PMC article. No abstract available. - A dynamic Boolean network reveals that the BMI1 and MALAT1 axis is associated with drug resistance by limiting miR-145-5p in non-small cell lung cancer.
Gupta S, Silveira DA, Piedade GPS, Ostrowski MP, Mombach JCM, Hashimoto RF. Gupta S, et al. Noncoding RNA Res. 2023 Oct 19;9(1):185-193. doi: 10.1016/j.ncrna.2023.10.008. eCollection 2024 Mar. Noncoding RNA Res. 2023. PMID: 38125755 Free PMC article. - MYC function and regulation in physiological perspective.
Jha RK, Kouzine F, Levens D. Jha RK, et al. Front Cell Dev Biol. 2023 Oct 24;11:1268275. doi: 10.3389/fcell.2023.1268275. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37941901 Free PMC article. Review. - Modular pooled discovery of synthetic knockin sequences to program durable cell therapies.
Blaeschke F, Chen YY, Apathy R, Daniel B, Chen AY, Chen PA, Sandor K, Zhang W, Li Z, Mowery CT, Yamamoto TN, Nyberg WA, To A, Yu R, Bueno R, Kim MC, Schmidt R, Goodman DB, Feuchtinger T, Eyquem J, Jimmie Ye C, Carnevale J, Satpathy AT, Shifrut E, Roth TL, Marson A. Blaeschke F, et al. Cell. 2023 Sep 14;186(19):4216-4234.e33. doi: 10.1016/j.cell.2023.08.013. Cell. 2023. PMID: 37714135 Free PMC article. - Squalene epoxidase/SQLE is a candidate target for treatment of colorectal cancers with p53 mutation and elevated c-MYC expression.
Du Y, Rokavec M, Hermeking H. Du Y, et al. Int J Biol Sci. 2023 Aug 6;19(13):4103-4122. doi: 10.7150/ijbs.85724. eCollection 2023. Int J Biol Sci. 2023. PMID: 37705742 Free PMC article.
References
- Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. - PubMed
- Meyer N, Kim SS, Penn LZ. The Oscar-worthy role of Myc in apoptosis. Semin Cancer Biol. 2006;16:275–287. - PubMed
- Dang CV, et al. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–264. - PubMed
- Berns K, Hijmans EM, Koh E, Daley GQ, Bernards R. A genetic screen to identify genes that rescue the slow growth phenotype of c-myc null fibroblasts. Oncogene. 2000;19:3330–3334. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous