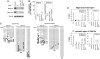G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription - PubMed (original) (raw)
G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription
Makoto Tachibana et al. EMBO J. 2008.
Abstract
Methylation of DNA and lysine 9 of histone H3 (H3K9) are well-conserved epigenetic marks for transcriptional silencing. Although H3K9 methylation directs DNA methylation in filamentous fungi and plants, this pathway has not been corroborated in mammals. G9a and GLP/Eu-HMTase1 are two-related mammalian lysine methyltransferases and a G9a/GLP heteromeric complex regulates H3K9 methylation of euchromatin. To elucidate the function of G9a/GLP-mediated H3K9 methylation in the regulation of DNA methylation and transcriptional silencing, we characterized ES cells expressing catalytically inactive mutants of G9a and/or GLP. Interestingly, in ES cells expressing a G9a-mutant/GLP complex that does not rescue global H3K9 methylation, G9a/GLP-target genes remain silent. The CpG sites of the promoter regions of these genes were hypermethylated in such mutant ES cells, but hypomethylated in G9a- or GLP-KO ES cells. Treatment with a DNA methyltransferase inhibitor reactivates these G9a/GLP-target genes in ES cells expressing catalytically inactive G9a/GLP proteins, but not the wild-type proteins. This is the first clear evidence that G9a/GLP suppresses transcription by independently inducing both H3K9 and DNA methylation.
Figures
Figure 1
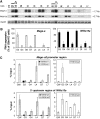
Reconstitution of G9a/GLP complex carrying mutations in their SET-domains. (A) Amino-acid alignment of the SET-domains of mG9a-L (Genbank, 21832048) and mGLP (Genbank, 60115440). Conserved and similar residues (shown as +) are shown between the two sequences. Mutagenized amino acids are shown in grey boxes. (B) Table showing serial mutants of G9a and GLP. (C) In vitro histone methyltransferase activity of mutant SET-domains of G9a (left) and GLP (right). GST-fused carboxy terminus of mouse G9a (969-1263 of G9a-L) and that of mouse GLP (1002–1296) were incubated with recombinant histone H3/H4 and _S_-adenosyl-[_methyl_-14C]-
L
-methionine, and then separated by SDS–PAGE. Coomassie staining shows enzymes and substrates. Autoradiography shows 14C-methyl group incorporated into recombinant H3. Activities of mutant enzymes were quantified using image J (
) and those of GWT or LWT are normalized to 100%. (D) List of ES cell lines expressing mutant G9a/GLP complexes. Serial cell lines were established by stable introduction of the expression plasmids for full-length of G9a-L or GLP into the corresponding KO cells shown in Supplementary Figure S1. Expression levels of mutant G9a or GLP proteins are shown in Supplementary Figure S2. (E) The ability of serial mutant proteins to form heterodimers are numerically represented as G9a/GLP interaction index. Protein levels within anti-G9a or anti-GLP immune complexes shown in Supplementary Figure S3 were measured by image J software, and the ratio of GLP protein immunoprecipitated with anti-G9a to those immunoprecipitated with anti-GLP were calculated. The interaction index of wild-type (WT) ES cells (top left panel) is ∼100%, indicating that endogenous G9a and GLP form a stoichiometric (1:1) heteromer in wild-type cells. (F) H3K9 methylation status of serial mutant ES lines were measured by immunoblot analysis and numerically represented as H3K9me recovery (Raw data are presented in Supplementary Figure S4). The values of signal intensities for methylated H3K9 of GW and LW are normalized to 100%, and those of the corresponding KO cells are normalized to 0%. Data represents an average value of the H3K9 recovery of two clones established independently.
Figure 2
Effect of G9a or GLP mutations on transcriptional suppression. (A) Northern blot analyses of Mage-a and Wfdc15a genes in ES cells expressing mutant G9a/GLP are shown. Total RNA collected from two independent clones were separated and probed with radiolabelled cDNAs. (B) Signal intensities for the transcripts of Mage-a (left) and Wfdc15a (right) were calculated by image J software and numerically represented. The value for _G9a-_KO cells was set at 100%. Data represents an average value between two independent clones. (C) Differences in the histone modification patterns on the Mage-a2 promoter region (top) and 5′-upstream region of Wfdc15a (bottom) in the serial ES cell lines were measured by chromatin immunoprecipitation (ChIP) analysis using anti-H3K9me1 and me2 (left) and H3K4me2 (right). Each experiment was performed twice.
Figure 3
G9a/GLP complex formation induces DNA methylation of the Mage-a2 promoter and the 5′-upstream region of the Wfdc15a gene. (A) A schematic diagram of CpG positions and amplicon for ChIP analysis within Mage-a2 promoter is shown. DNA methylation status of eight CpG sequences (no. 1-8) within the promoter (0 to −287) and a CpG (no. 9) located at the 5′ end of the transcribed region were examined in this study. (B) Methylation differences among serial ES lines expressing mutant G9a/GLP complex at the Mage-a2 promoter. DNA methylation status of CpG positions nos. 1–9 shown in (A) was measured by bisulphite sequencing. Open circles indicate unmethylated cytosines and filled circles indicate methylated cytosines. (C) Schematic diagram of CpG positions and amplicon for ChIP analysis within the 5′-upstream region of Wfdc15a. (D, E) Methylation differences among serial ES lines expressing mutant G9a/GLP complex at the position no. 1–8 (D) and no. 9–11 (E) of CpG sequences on the 5′-upstream region of Wfdc15a.
Figure 4
Inhibition of DNA methyltransferase activity reactivates G9a/GLP-target gene expression independent of H3K9 methylation. (A) Serial ES lines were cultured with histone deacetylase inhibitor, tricostatin-A (TSA) or the DNA methyltransferase inhibitor, 5-Aza-2′-deoxycytidine (5AZ). Total RNA from treated cells was isolated and analysed through northern blotting using radiolabelled cDNA. (B) The effect of TSA on H3 acetylation was confirmed by immunoblot analysis with specific antibodies directed against acetyl histone H3. Ponceau staining represents equal loading of histones. (C) Reactivation of the transcripts for Mage-a (left) and Wfdc15a (right) is represented numerically. The values for _G9a-_KO cells without drug treatment are normalized to 100%. Experiments were performed twice.
Figure 5
H3K9 methylation suppresses G9a/GLP-target genes in DNA methyltransferase-deficient ES cells. (A) Northern blot analyses of _Dnmts-_KO ES cells are shown. Total RNA was separated and probed with radiolabelled cDNAs. (B) Expression levels for Mage-a (left) and Wfdc15a (right) are numerically represented. The values for _G9a-_KO cells are normalized to 100%. Each experiment was carried out twice. (C) Cytosine methylation status of Mage-a2 promoter and 5′-upstream region of Wfdc15a in _Dnmt1-_KO (left) and Dnmt3a+b DKO (right) ES cells. CpG positions are as described in Figure 3B and D. (D) Histone methylation patterns of the Mage-a2 promoter region (top) and 5′-upstream region of Wfdc15a (bottom) in _Dnmts-_KO ES cells were determined by ChIP analyses. H3K9me2 around the promoter is still maintained in _Dnmt1-_KO and Dnmt3a+b DKO ES cells. Experiments were carried out twice.
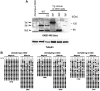
Figure 6
Dnmt3a and 3b redundantly methylate CpG sequences on G9a/GLP-target genes. (A) Dnmt3 protein expression of several rescued ES lines on Dnmt3a+b DKO background, were analysed with mouse monoclonal antibody 64B1446, which recognizes carboxy terminus of both Dnmt3a and 3b (Chen et al, 2002). (B) Cytosine methylation status of Mage-a2 promoter and 5′-upstream region of Wfdc15a in Dnmt3-rescued line. CpG positions are as described in Figure 3B and D.
Figure 7
Schematic representation of G9a/GLP complex-mediated gene suppression. One candidate molecule links G9a/GLP to DNA methyltransferase is Wiz, as it binds preferentially G9a/GLP heteromer (Ueda et al, 2006). Genetic analyses indicate that orphan G9a (right) or GLP (left) is both incapable of catalysing H3K9 methylation and recruiting DNA methylation, accompanied with the reactivation of the target genes (see panels of _G9a-_KO and _GLP_-KO). Mutant complex incapable of H3K9 methylation, such as GM4/LWT or GM7/LWT, can still suppress transcription via recruiting DNA methyltransferase(s) (see panels of inactive G9a/GLP). This suppression is cancelled by 5′Aza-dC treatment. G9a/GLP complex can also suppress transcription even without DNA methylation, solely by depositing H3K9 methylation (see panel of _Dnmts-_KO).
Similar articles
- DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity.
Dong KB, Maksakova IA, Mohn F, Leung D, Appanah R, Lee S, Yang HW, Lam LL, Mager DL, Schübeler D, Tachibana M, Shinkai Y, Lorincz MC. Dong KB, et al. EMBO J. 2008 Oct 22;27(20):2691-701. doi: 10.1038/emboj.2008.193. Epub 2008 Sep 25. EMBO J. 2008. PMID: 18818693 Free PMC article. - Zinc finger protein Wiz links G9a/GLP histone methyltransferases to the co-repressor molecule CtBP.
Ueda J, Tachibana M, Ikura T, Shinkai Y. Ueda J, et al. J Biol Chem. 2006 Jul 21;281(29):20120-8. doi: 10.1074/jbc.M603087200. Epub 2006 May 15. J Biol Chem. 2006. PMID: 16702210 - Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9.
Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y. Tachibana M, et al. Genes Dev. 2005 Apr 1;19(7):815-26. doi: 10.1101/gad.1284005. Epub 2005 Mar 17. Genes Dev. 2005. PMID: 15774718 Free PMC article. - Regulation of cell differentiation and function by the euchromatin histone methyltranserfases G9a and GLP.
Kramer JM. Kramer JM. Biochem Cell Biol. 2016 Feb;94(1):26-32. doi: 10.1139/bcb-2015-0017. Epub 2015 Jun 1. Biochem Cell Biol. 2016. PMID: 26198080 Review. - Structure and function of histone H3 lysine 9 methyltransferases and demethylases.
Krishnan S, Horowitz S, Trievel RC. Krishnan S, et al. Chembiochem. 2011 Jan 24;12(2):254-63. doi: 10.1002/cbic.201000545. Epub 2011 Jan 5. Chembiochem. 2011. PMID: 21243713 Review.
Cited by
- Breaking through an epigenetic wall: re-activation of Oct4 by KRAB-containing designer zinc finger transcription factors.
Juárez-Moreno K, Erices R, Beltran AS, Stolzenburg S, Cuello-Fredes M, Owen GI, Qian H, Blancafort P. Juárez-Moreno K, et al. Epigenetics. 2013 Feb;8(2):164-76. doi: 10.4161/epi.23503. Epub 2013 Jan 11. Epigenetics. 2013. PMID: 23314702 Free PMC article. - Epigenetics of the failing heart.
Marín-García J, Akhmedov AT. Marín-García J, et al. Heart Fail Rev. 2015 Jul;20(4):435-59. doi: 10.1007/s10741-015-9483-x. Heart Fail Rev. 2015. PMID: 25847519 Review. - Methionine adenosyltransferase II-dependent histone H3K9 methylation at the COX-2 gene locus.
Kera Y, Katoh Y, Ohta M, Matsumoto M, Takano-Yamamoto T, Igarashi K. Kera Y, et al. J Biol Chem. 2013 May 10;288(19):13592-601. doi: 10.1074/jbc.M112.429738. Epub 2013 Mar 28. J Biol Chem. 2013. PMID: 23539621 Free PMC article. - The Emerging Role of ATP-Dependent Chromatin Remodeling in Memory and Substance Use Disorders.
López AJ, Hecking JK, White AO. López AJ, et al. Int J Mol Sci. 2020 Sep 17;21(18):6816. doi: 10.3390/ijms21186816. Int J Mol Sci. 2020. PMID: 32957495 Free PMC article. Review. - EHMT1 protein binds to nuclear factor-κB p50 and represses gene expression.
Ea CK, Hao S, Yeo KS, Baltimore D. Ea CK, et al. J Biol Chem. 2012 Sep 7;287(37):31207-17. doi: 10.1074/jbc.M112.365601. Epub 2012 Jul 16. J Biol Chem. 2012. PMID: 22801426 Free PMC article.
References
- Bannister A, Zegerman P, Partridge J, Miska E, Thomas J, Allshire R, Kouzarides T (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124 - PubMed
- Bird A (2002) DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21 - PubMed
- Bostick M, Kim J, Estève P, Clark A, Pradhan S, Jacobsen S (2007) UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317: 1760–1764 - PubMed
- Chen T, Ueda Y, Xie S, Li E (2002) A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J Biol Chem 277: 38746–38754 - PubMed
- De Plaen E, De Backer O, Arnaud D, Bonjean B, Chomez P, Martelange V, Avner P, Baldacci P, Babinet C, Hwang S, Knowles B, Boon T (1999) A new family of mouse genes homologous to the human MAGE genes. Genomics 55: 176–184 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials