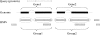GenBlastA: enabling BLAST to identify homologous gene sequences - PubMed (original) (raw)
GenBlastA: enabling BLAST to identify homologous gene sequences
Rong She et al. Genome Res. 2009 Jan.
Abstract
BLAST is an extensively used local similarity search tool for identifying homologous sequences. When a gene sequence (either protein sequence or nucleotide sequence) is used as a query to search for homologous sequences in a genome, the search results, represented as a list of high-scoring pairs (HSPs), are fragments of candidate genes rather than full-length candidate genes. Relevant HSPs ("signals"), which represent candidate genes in the target genome sequences, are buried within a report that contains also hundreds to thousands of random HSPs ("noises"). Consequently, BLAST results are often overwhelming and confusing even to experienced users. For effective use of BLAST, a program is needed for extracting relevant HSPs that represent candidate homologous genes from the entire HSP report. To achieve this goal, we have designed a graph-based algorithm, genBlastA, which automatically filters HSPs into well-defined groups, each representing a candidate gene in the target genome. The novelty of genBlastA is an edge length metric that reflects a set of biologically motivated requirements so that each shortest path corresponds to an HSP group representing a homologous gene. We have demonstrated that this novel algorithm is both efficient and accurate for identifying homologous sequences, and that it outperforms existing approaches with similar functionalities.
Figures
Figure 1.
Grouping of HSPs into groups representing paralogs (Gene1 and Gene2) in tandem in the target genome. For simplicity, this figure shows only a small portion of the HSPs returned by BLAST. Each HSP may correspond to a coding segment (likely an exon) of a gene, thus a group of HSPs may collectively represent a full-length gene. Each shaded box at the bottom of the figures represents an HSP at its corresponding genomic position. Candidate genes are shown on the genome, with exons (black boxes) connected by introns (lines). The HSP groups that best represent the genes are shown under the corresponding genes, with relevant HSPs in the groups circled. Two paralogous genes in tandem (Gene1 and Gene2) are shown. The boundary of the two genes must be correctly resolved.
Figure 2.
Grouping HSPs into groups representing individual genes. genBlastA was able to resolve all five members, while ML resolved only two and WU only one. Gene models are shown in the Gene Models track. HSPs are shown as blue boxes in the All HSPs track. The color indicates different PIDs for the HSPs. Darker color indicates higher PID. The genBlastA Group, ML Group, and WU Group tracks show HSPs groupings that are returned by genBlastA, ML, and WU-BLAST, respectively.
Figure 3.
Grouping of HSPs to represent individual homologous genes in tandem clusters. This figure shows average resolve rate for a total of 30 tandem duplicated gene clusters in the EvsE data set for genBlastA (GB), Cui et al. (2007) (ML), and WU-Blast (WU). Ratio of specific groups was calculated as the number of genes resolved over the total number of genes in each tandem gene cluster. A gene is considered resolved if the HSP group overlaps with only one single gene in WormBase and the span similarity is ≥50%. Gapped and ungapped represent two independent BLAST results using either gapped setting or ungapped setting. GB alpha value is 0.5. ML distance threshold is 1000. Error bars, SE. (***) Statistical significance (P < 0.001) by paired Student’s _t_-test.
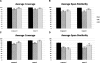
Figure 4.
(A) Average coverage for EvsE data set. (B) Average span similarity for EvsE data set. (C) Average coverage for EvsB data set. (D) Average span similarity for EvsB data set. In all cases, figures represent averaged results over 464 test genes for three different programs genBlastA (GB), Cui et al. (2007) (ML), and WU-Blast (WU). Gapped and ungapped represent two independent BLAST results using either gapped setting or ungapped setting. Span similarity is calculated by Jaccard similarity. GB alpha value is 0.5. ML distance threshold is 1000. Error bars, SE. (***) Statistical significance (P < 0.001) by paired Student’s _t_-test.
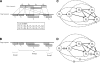
Figure 5.
(A) HSPs returned by BLAST. Q1, Q2, Q3, and Q4 represent query segments, while T1, T2, T3, T4, T5, and T6 represent target segments. (B) Example of groups of HSPs. (C) The HSP graph, with solid lines representing edges and dotted edges indicating skip edges. (D) The HSP graph, with vertical bars indicating separating edges.
Similar articles
- Homology search for genes.
Cui X, Vinar T, Brejová B, Shasha D, Li M. Cui X, et al. Bioinformatics. 2007 Jul 1;23(13):i97-103. doi: 10.1093/bioinformatics/btm225. Bioinformatics. 2007. PMID: 17646351 - genBlastG: using BLAST searches to build homologous gene models.
She R, Chu JS, Uyar B, Wang J, Wang K, Chen N. She R, et al. Bioinformatics. 2011 Aug 1;27(15):2141-3. doi: 10.1093/bioinformatics/btr342. Epub 2011 Jun 8. Bioinformatics. 2011. PMID: 21653517 - MAP: searching large genome databases.
Kahveci T, Singh A. Kahveci T, et al. Pac Symp Biocomput. 2003:303-14. Pac Symp Biocomput. 2003. PMID: 12603037 - TruMatch--a BLAST post-processor that identifies bona fide sequence matches to genome assemblies.
Li W, Rehmeyer CJ, Staben C, Farman ML. Li W, et al. Bioinformatics. 2005 May 1;21(9):2097-8. doi: 10.1093/bioinformatics/bti257. Epub 2005 Jan 25. Bioinformatics. 2005. PMID: 15671115 - Tracembler--software for in-silico chromosome walking in unassembled genomes.
Dong Q, Wilkerson MD, Brendel V. Dong Q, et al. BMC Bioinformatics. 2007 May 9;8:151. doi: 10.1186/1471-2105-8-151. BMC Bioinformatics. 2007. PMID: 17490482 Free PMC article.
Cited by
- Whole-Genome Sequencing of the Entomopathogenic Fungus Fusarium solani KMZW-1 and Its Efficacy Against Bactrocera dorsalis.
Yu J, Hussain M, Wu M, Shi C, Li S, Ji Y, Hussain S, Qin D, Xiao C, Wu G. Yu J, et al. Curr Issues Mol Biol. 2024 Oct 17;46(10):11593-11612. doi: 10.3390/cimb46100688. Curr Issues Mol Biol. 2024. PMID: 39451568 Free PMC article. - Development of genomic and genetic resources facilitating molecular genetic studies on untapped Myanmar rice germplasms.
Furuta T, Saw OM, Moe S, Win KT, Hlaing MM, Hlaing ALL, Thein MS, Yasui H, Ashikari M, Yoshimura A, Yamagata Y. Furuta T, et al. Breed Sci. 2024 Apr;74(2):124-137. doi: 10.1270/jsbbs.23077. Epub 2024 Mar 22. Breed Sci. 2024. PMID: 39355624 Free PMC article. - Genome-Wide Identification and Expression Analysis of MYB Transcription Factor Family in Response to Various Abiotic Stresses in Coconut (Cocos nucifera L.).
Si CC, Li YB, Hai X, Bao CC, Zhao JY, Ahmad R, Li J, Wang SC, Li Y, Yang YD. Si CC, et al. Int J Mol Sci. 2024 Sep 18;25(18):10048. doi: 10.3390/ijms251810048. Int J Mol Sci. 2024. PMID: 39337532 Free PMC article. - The Chromosome-level Genome Provides Insights into the Evolution and Adaptation of Extreme Aggression.
Liu PC, Wang ZY, Qi M, Hu HY. Liu PC, et al. Mol Biol Evol. 2024 Sep 4;41(9):msae195. doi: 10.1093/molbev/msae195. Mol Biol Evol. 2024. PMID: 39271164 Free PMC article. - Chromosome-level genome assembly of Oriental chestnut gall wasp (Dryocosmus kuriphilus).
Liu B, Ren YS, Su CY, Wang XD, Zeng Y, Zhu DH. Liu B, et al. Sci Data. 2024 Sep 4;11(1):963. doi: 10.1038/s41597-024-03827-7. Sci Data. 2024. PMID: 39232034 Free PMC article.
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
- Bentley D.R. Whole-genome re-sequencing. Curr. Opin. Genet. Dev. 2006;16:545–552. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials