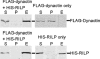Huntingtin regulates RE1-silencing transcription factor/neuron-restrictive silencer factor (REST/NRSF) nuclear trafficking indirectly through a complex with REST/NRSF-interacting LIM domain protein (RILP) and dynactin p150 Glued - PubMed (original) (raw)
Huntingtin regulates RE1-silencing transcription factor/neuron-restrictive silencer factor (REST/NRSF) nuclear trafficking indirectly through a complex with REST/NRSF-interacting LIM domain protein (RILP) and dynactin p150 Glued
Masahito Shimojo. J Biol Chem. 2008.
Abstract
Huntingtin has been reported to regulate the nuclear translocation of the transcriptional repressor RE1-silencing transcription factor/neuron-restrictive silencer factor (REST/NRSF). The REST/NRSF-interacting LIM domain protein (RILP) has also been shown to regulate REST/NRSF nuclear translocation. Therefore, we were prompted to address the question of how two distinct proteins could have the same function. We initially used a yeast two-hybrid screen to look for an interaction between huntingtin and RILP. This screen identified dynactin p150(Glued) as an interacting protein. Coimmunoprecipitation of proteins in vitro expressed in a reticulocyte lysate system showed an interaction between REST/NRSF and RILP as well as between RILP and dynactin p150(Glued). Coimmunoprecipitation analysis further showed a complex containing RILP, dynactin p150(Glued), and huntingtin. Huntingtin did not interact directly with either REST/NRSF or RILP, but did interact with dynactin p150(Glued). The N-terminal fragment of wild-type huntingtin did not affect the interaction between dynactin p150(Glued) and RILP; however, mutant huntingtin weakened this interaction. We further show that HAP1 (huntingtin-associated protein-1) prevents this complex from translocating REST/NRSF to the nucleus. Thus, this study suggests that REST/NRSF, dynactin p150(Glued), huntingtin, HAP1, and RILP form a complex involved in the translocation of REST/NRSF into the nucleus and that HAP1 controls REST/NRSF cellular localization in neurons.
Figures
FIGURE 1.
Pulldown assays for analyzing the interaction of RILP and dynactin p150Glued. His-tagged RILP and FLAG-tagged dynactin p150Glued were coexpressed in the TNT coupled reticulocyte lysate system. The lysate was applied to a Ni-NTA-agarose column, and the various fractions were subjected to SDS-PAGE, followed by Western blot analysis using anti-FLAG (upper panels) or anti-His (lower panels) antibody. S, starting lysate; P, passthrough; E, eluate.
FIGURE 2.
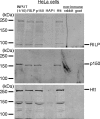
Coimmunoprecipitation of RILP with dynactin p150Glued and huntingtin from HeLa cells. HeLa cell lysates were prepared in lysis buffer and subjected to immunoprecipitation with anti-RILP, anti-dynactin p150Glued (p150), anti-huntingtin (Htt), or nonimmune antiserum, respectively. Nonimmune antibodies were from goat and rabbit. Aliquots of immunoprecipitates were subjected to Western blot analysis with anti-RILP, anti-dynactin p150Glued, or anti-huntingtin antibodies. INPUT is one-tenth of what was applied to immunoprecipitation.
FIGURE 3.
Analysis of the direct interaction of huntingtin and REST/NRSF. Proteins were coexpressed in the TNT coupled reticulocyte lysate system. Lysates were subjected to immunoprecipitation (IP) with anti-RILP (α_-RILP_), anti-dynactin p150Glued (α_-p150_), anti-huntingtin (α_-Htt_), or anti-NRSF (α_-NRSF_) antibodies. Aliquots were subjected to Western blot analysis (5% SDS-polyacrylamide gel) with the indicated antibodies. + and -, presence and absence, respectively, of each protein in the lysate; S, starting lysate for immunoprecipitation; wt, wild-type; mut, mutant. The input was one-fifth of what was applied to immunoprecipitation.
FIGURE 4.
RILP interacts with huntingtin through dynactin p150Glued. Each protein was coexpressed and subjected to immunoprecipitation (IP) using the appropriate antibodies as shown in Fig. 3. Immunoprecipitation in the absence (a) or presence (b) of dynactin p150Glued was carried out with anti-RILP (α_-RILP_), anti-dynactin p150Glued (α_-p150_), or anti-huntingtin (α_-Htt_) antibodies. Aliquots of immunoprecipitates were subjected to Western blot analysis with anti-RILP, anti-p150, or anti-Htt antibodies. + and -, presence and absence, respectively, of each protein in the lysate; S, starting lysate for immunoprecipitation;wild, wild-type; mut, mutant. The input was one-fifth of what was applied to immunoprecipitation.
FIGURE 5.
Effect of HAP1 on the interaction between RILP and huntingtin. a, RILP coimmunoprecipitated with dynactin p150Glued, HAP1, and huntingtin in NT2 cells. NT2 cell lysates were prepared in lysis buffer and subjected to immunoprecipitation (IP) with anti-RILP, anti-dynactin p150Glued (p150), anti-huntingtin (Htt), anti-HAP1, or nonimmune antiserum. Nonimmune antibodies were from goat and rabbit. Aliquots of immunoprecipitates were subjected to Western blot analysis with antibodies. INPUT is one-tenth what was applied to immunoprecipitation. b, proteins were coexpressed and subjected to immunoprecipitation using anti-huntingtin or anti-RILP antibodies as described in the legend to Fig. 3. Immunoprecipitation was carried out with anti-RILP (α_-RILP_) or anti-huntingtin (α_-Htt_) antibodies. Aliquots of immunoprecipitates were subjected to Western blot analysis with anti-RILP or anti-Htt antibodies. + and -, presence and absence, respectively, of each protein in the lysate; S, starting lysate for immunoprecipitation. The input was one-fifth of what was applied to immunoprecipitation.c, the interaction between REST/NRSF and HAP1 or RILP and HAP1 was analyzed. Immunoprecipitation was carried out with anti-HAP1 (α_-HAP_), anti-RILP, or anti-NRSF (α_-NRSF_) antibodies. Aliquots of immunoprecipitates were subjected to Western blot analysis with anti-RILP, anti-NRSF, or anti-HAP1 antibodies.
FIGURE 6.
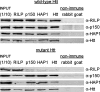
Coimmunoprecipitation of complexes from mouse striatal cells expressing wild-type or mutant huntingtin. Cell lysates were prepared in lysis buffer and subjected to immunoprecipitation with anti-RILP, anti-dynactin p150Glued (p150), anti-huntingtin (Htt), or nonimmune antiserum. Nonimmune antibodies were from goat and rabbit. Aliquots of immunoprecipitates were subjected to Western blot analysis with anti-RILP (α_-RILP_), anti-dynactin p150Glued (α_-p150_), or anti-huntingtin (α_-Htt_) antibodies.INPUT is one-tenth of what was applied to immunoprecipitation.
FIGURE 7.
Effect of HAP1 on the expression of reporter gene constructs containing an RE1/NRSE element in HeLa and NT2 cells. Reporter gene assays were performed using the 5′-noncoding region of the human choline acetyltransferase gene driving the luciferase reporter gene. Reporter constructs were pXP2-EX, which contains a cholinergic gene locus fragment with an active RE1/NRSE, and pXP2-EXmut, which contains a mutant inactive RE1/NRSE. Constructs were transiently transfected into the indicated cell lines as described under “Experimental Procedures.” Cells were transfected with HAP1 expression plasmid (+HAP1) or siRNA for HAP1. For the control, vehicle pcDNA3.1 plasmid or a scrambled siRNA was also transfected. After 24 h, reporter gene constructs with β-galactosidase plasmid were transfected with Effectene reagent. After transfection, cells were harvested, and extracts were prepared for assays of luciferase activity.a, luciferase activity is reported as the relative reporter activity normalized to β-galactosidase activity. Data are the means ± S.E. (n = 6). b, each lysate expressing HAP1 was subjected to SDS-PAGE, followed by Western blot analysis using anti-HAP1 or anti-tubulin antibodies.
FIGURE 8.
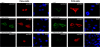
Effect of HAP1 expression on REST/NRSF localization. HeLa and NT2 cells were grown on a coverslip and transfected with HAP1 siRNA or HAP1 expression vector. After 24 h of culture, REST/NRSF was transfected, followed by culturing for an additional 24 h. The cells were then fixed with dry ice/cold methanol and incubated with anti-REST/NRSF (12C11-1) and anti-HAP1 antibodies followed by fluorescently labeled IgG. Red fluorescence is shown as REST/NRSF; green fluorescence is due to RILP; and_blue_ fluorescence is due to nuclear 4′,6-diamidino-2-phenylindole (DAPI) staining. Upper panels, cells transfected with REST/NRSF as well as with a non-related sequence control siRNA; middle panels, cells treated with HAP1 following transfection with REST/NRSF; lower panels, cells transfected with HAP1 siRNA followed transfecting with REST/NRSF. The subcellular distribution of REST was examined by confocal microscopy as described under “Experimental Procedures.” The relative staining intensities yielded the following percentages of nuclear REST in HeLa cells: 91.24 ± 3.17% (+REST), 20.19 ± 4.42% (+REST and HAP1), and 91.86 ± 1.70% (+REST and HAP1 siRNA). The percentages of nuclear REST in NT2 cells were as follows: 9.73 ± 2.09% (+REST), 9.71 ± 2.20% (+REST and HAP1), and 81.73 ± 2.92% (+REST and HAP1 siRNA).
Similar articles
- Characterization of the REST/NRSF-interacting LIM domain protein (RILP): localization and interaction with REST/NRSF.
Shimojo M, Hersh LB. Shimojo M, et al. J Neurochem. 2006 Feb;96(4):1130-8. doi: 10.1111/j.1471-4159.2005.03608.x. Epub 2006 Jan 17. J Neurochem. 2006. PMID: 16417580 - Huntingtin-associated protein 1 (HAP1) interacts with the p150Glued subunit of dynactin.
Engelender S, Sharp AH, Colomer V, Tokito MK, Lanahan A, Worley P, Holzbaur EL, Ross CA. Engelender S, et al. Hum Mol Genet. 1997 Dec;6(13):2205-12. doi: 10.1093/hmg/6.13.2205. Hum Mol Genet. 1997. PMID: 9361024 - REST/NRSF-interacting LIM domain protein, a putative nuclear translocation receptor.
Shimojo M, Hersh LB. Shimojo M, et al. Mol Cell Biol. 2003 Dec;23(24):9025-31. doi: 10.1128/MCB.23.24.9025-9031.2003. Mol Cell Biol. 2003. PMID: 14645515 Free PMC article. - [Huntington's disease: intracellular signaling pathways and neuronal death].
Humbert S, Saudou F. Humbert S, et al. J Soc Biol. 2005;199(3):247-51. doi: 10.1051/jbio:2005026. J Soc Biol. 2005. PMID: 16471265 Review. French. - Regulation of the cholinergic gene locus by the repressor element-1 silencing transcription factor/neuron restrictive silencer factor (REST/NRSF).
Shimojo M, Hersh LB. Shimojo M, et al. Life Sci. 2004 Mar 19;74(18):2213-25. doi: 10.1016/j.lfs.2003.08.045. Life Sci. 2004. PMID: 15017977 Review.
Cited by
- Long Non-coding RNAs, Novel Culprits, or Bodyguards in Neurodegenerative Diseases.
Wang DQ, Fu P, Yao C, Zhu LS, Hou TY, Chen JG, Lu Y, Liu D, Zhu LQ. Wang DQ, et al. Mol Ther Nucleic Acids. 2018 Mar 2;10:269-276. doi: 10.1016/j.omtn.2017.12.011. Epub 2017 Dec 22. Mol Ther Nucleic Acids. 2018. PMID: 29499939 Free PMC article. - Huntingtin loss in hepatocytes is associated with altered metabolism, adhesion, and liver zonation.
Bragg RM, Coffey SR, Cantle JP, Hu S, Singh S, Legg SR, McHugh CA, Toor A, Zeitlin SO, Kwak S, Howland D, Vogt TF, Monga SP, Carroll JB. Bragg RM, et al. Life Sci Alliance. 2023 Sep 8;6(11):e202302098. doi: 10.26508/lsa.202302098. Print 2023 Nov. Life Sci Alliance. 2023. PMID: 37684045 Free PMC article. - Navigating the neuronal recycling bin: Another look at huntingtin in coordinating autophagy.
Krzystek TJ, Gunawardena S. Krzystek TJ, et al. Autophagy Rep. 2025 Jun 2;4(1):2472450. doi: 10.1080/27694127.2025.2472450. eCollection 2025. Autophagy Rep. 2025. PMID: 40475846 Free PMC article. Review. - Huntingtin-associated protein 1-associated intracellular trafficking in neurodegenerative diseases.
Chen X, He E, Su C, Zeng Y, Xu J. Chen X, et al. Front Aging Neurosci. 2023 Feb 7;15:1100395. doi: 10.3389/fnagi.2023.1100395. eCollection 2023. Front Aging Neurosci. 2023. PMID: 36824265 Free PMC article. Review. - The interaction between RE1-silencing transcription factor (REST) and heat shock protein 90 as new therapeutic target against Huntington's disease.
Orozco-Díaz R, Sánchez-Álvarez A, Hernández-Hernández JM, Tapia-Ramírez J. Orozco-Díaz R, et al. PLoS One. 2019 Jul 30;14(7):e0220393. doi: 10.1371/journal.pone.0220393. eCollection 2019. PLoS One. 2019. PMID: 31361762 Free PMC article.
References
- Harper, P. S. (ed) (1991) Huntington's Disease, W. B. Saunders Co., Philadelphia, PA
- Bates, G., Harper, P. S., and Jones, L. (eds) (2002) Huntington's Disease, Oxford University Press, Oxford
- Zeitlin, S., Liu, J. P., Chapman, D. L., Papaioannou, V. E., and Efstratiadis, A. (1995) Nat. Genet. 11 155-163 - PubMed
- Nasir, J., Floresco, S. B., O'Kusky, J. R., Diewert, V. M., Richman, J. M., Zeisler, J., Borowski, A., Marth, J. D., Phillips, A. G., and Hayden, M. R. (1995) Cell 81 811-823 - PubMed
- Huntington's Disease Collaborative Research Group (1993) Cell 72 971-983 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous