Rfam: updates to the RNA families database - PubMed (original) (raw)
. 2009 Jan;37(Database issue):D136-40.
doi: 10.1093/nar/gkn766. Epub 2008 Oct 25.
Affiliations
- PMID: 18953034
- PMCID: PMC2686503
- DOI: 10.1093/nar/gkn766
Rfam: updates to the RNA families database
Paul P Gardner et al. Nucleic Acids Res. 2009 Jan.
Abstract
Rfam is a collection of RNA sequence families, represented by multiple sequence alignments and covariance models (CMs). The primary aim of Rfam is to annotate new members of known RNA families on nucleotide sequences, particularly complete genomes, using sensitive BLAST filters in combination with CMs. A minority of families with a very broad taxonomic range (e.g. tRNA and rRNA) provide the majority of the sequence annotations, whilst the majority of Rfam families (e.g. snoRNAs and miRNAs) have a limited taxonomic range and provide a limited number of annotations. Recent improvements to the website, methodologies and data used by Rfam are discussed. Rfam is freely available on the Web at http://rfam.sanger.ac.uk/and http://rfam.janelia.org/.
Figures
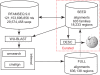
Figure 1.
An outline of the Rfam 9.0 databases and methods. RFAMSEQ is drawn from EMBL excluding only the EST, synthetic and patented divisions. There are 603 Rfam families in release 9.0, which are used to scan RFAMSEQ for homologues using first WU-BLAST filters followed by the more accurate CM-based methods cmsearch and cmalign. This results in 603 FULL alignments annotating 636 138 regions.
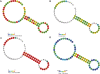
Figure 2.
An example of the new secondary markups used by Rfam. The coronavirus 3′-UTR pseudoknot is shown (Rfam Accession RF00165). We display coloured markups of sequence conservation (A), covariation (B), base-pair conservation also known as the fraction of canonical base pairs (C) and CM scores (D).
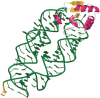
Figure 3.
An example of how PDB structures are displayed in Rfam. In this case, the structure 1l ng, containing the SRP19-7S.S RNA Complex from M. jannaschii, is rendered as cartoons using Jmol. Protein regions are coloured using the following scheme: beta-sheets (yellow), helices (magenta) and unstructured regions (white). RNA bases that match the Rfam model are coloured according to the key given in the web page (not shown here). In this structure, green represents a match to the eukaryotic SRP model, whereas those unmatched bases are coloured orange.
Similar articles
- Rfam 11.0: 10 years of RNA families.
Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP, Eddy SR, Gardner PP, Bateman A. Burge SW, et al. Nucleic Acids Res. 2013 Jan;41(Database issue):D226-32. doi: 10.1093/nar/gks1005. Epub 2012 Nov 3. Nucleic Acids Res. 2013. PMID: 23125362 Free PMC article. - Rfam: an RNA family database.
Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Griffiths-Jones S, et al. Nucleic Acids Res. 2003 Jan 1;31(1):439-41. doi: 10.1093/nar/gkg006. Nucleic Acids Res. 2003. PMID: 12520045 Free PMC article. - Rfam: annotating non-coding RNAs in complete genomes.
Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Griffiths-Jones S, et al. Nucleic Acids Res. 2005 Jan 1;33(Database issue):D121-4. doi: 10.1093/nar/gki081. Nucleic Acids Res. 2005. PMID: 15608160 Free PMC article. - Computational identification of functional RNA homologs in metagenomic data.
Nawrocki EP, Eddy SR. Nawrocki EP, et al. RNA Biol. 2013 Jul;10(7):1170-9. doi: 10.4161/rna.25038. Epub 2013 May 20. RNA Biol. 2013. PMID: 23722291 Free PMC article. Review. - miRBase: the microRNA sequence database.
Griffiths-Jones S. Griffiths-Jones S. Methods Mol Biol. 2006;342:129-38. doi: 10.1385/1-59745-123-1:129. Methods Mol Biol. 2006. PMID: 16957372 Review.
Cited by
- The full-length transcriptional of the multiple spatiotemporal embryo-gonad tissues in chicken (Gallus gallus).
Jin K, Zuo Q, Song J, Elsayed AK, Sun H, Niu Y, Zhang Y, Chang G, Chen G, Li B. Jin K, et al. BMC Genom Data. 2024 Oct 25;25(1):91. doi: 10.1186/s12863-024-01273-3. BMC Genom Data. 2024. PMID: 39455914 Free PMC article. - RAFFT: Efficient prediction of RNA folding pathways using the fast Fourier transform.
Opuu V, Merleau NSC, Messow V, Smerlak M. Opuu V, et al. PLoS Comput Biol. 2022 Aug 26;18(8):e1010448. doi: 10.1371/journal.pcbi.1010448. eCollection 2022 Aug. PLoS Comput Biol. 2022. PMID: 36026505 Free PMC article. - Functional and Structural Characterization of FAU Gene/Protein from Marine Sponge Suberites domuncula.
Perina D, Korolija M, Hadžija MP, Grbeša I, Belužić R, Imešek M, Morrow C, Marjanović MP, Bakran-Petricioli T, Mikoč A, Ćetković H. Perina D, et al. Mar Drugs. 2015 Jul 7;13(7):4179-96. doi: 10.3390/md13074179. Mar Drugs. 2015. PMID: 26198235 Free PMC article. - A map of the SARS-CoV-2 RNA structurome.
Andrews RJ, O'Leary CA, Tompkins VS, Peterson JM, Haniff HS, Williams C, Disney MD, Moss WN. Andrews RJ, et al. NAR Genom Bioinform. 2021 May 22;3(2):lqab043. doi: 10.1093/nargab/lqab043. eCollection 2021 Jun. NAR Genom Bioinform. 2021. PMID: 34046592 Free PMC article. - Complete genome sequence of the giant virus OBP and comparative genome analysis of the diverse ΦKZ-related phages.
Cornelissen A, Hardies SC, Shaburova OV, Krylov VN, Mattheus W, Kropinski AM, Lavigne R. Cornelissen A, et al. J Virol. 2012 Feb;86(3):1844-52. doi: 10.1128/JVI.06330-11. Epub 2011 Nov 30. J Virol. 2012. PMID: 22130535 Free PMC article.
References
- Cochrane G, Akhtar R, Aldebert P, Althorpe N, Baldwin A, Bates K, Bhattacharyya S, Bonfield J, Bower L, Browne P, et al. Priorities for nucleotide trace, sequence and annotation data capture at the Ensembl Trace Archive and the EMBL Nucleotide Sequence Database. Nucleic Acids Res. 2008;36:D5–D12. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials