MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons - PubMed (original) (raw)
MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons
Armaz Aschrafi et al. J Neurosci. 2008.
Abstract
MicroRNAs (miRs) are evolutionarily conserved, noncoding RNA molecules of approximately 21 nt that regulate the expression of genes that are involved in various biological processes, such as cell proliferation and differentiation. Previously, we reported the presence of a heterogeneous population of mRNAs present in the axons and nerve terminals of primary sympathetic neurons to include the nuclear-encoded mitochondrial mRNA coding for COXIV. Sequence analysis of the 3'UTR of this mRNA revealed the presence of a putative binding site for miR-338, a brain-specific microRNA. Transfection of precursor miR-338 into the axons of primary sympathetic neurons decreases COXIV mRNA and protein levels and results in a decrease in mitochondrial activity, as measured by the reduction of ATP levels. Conversely, the transfection of synthetic anti-miR oligonucleotides that inhibit miR-338 increases COXIV levels, and results in a significant increase in oxidative phosphorylation and also norepinephrine uptake in the axons. Our results point to a molecular mechanism by which this microRNA participates in the regulation of axonal respiration and function by modulating the levels of COXIV, a protein which plays a key role in the assembly of the mitochondrial cytochrome c oxidase complex IV.
Figures
Figure 1.
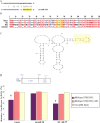
MiR-338 targets rat COXIV. A, Putative miR-338 binding site in the rat COXIV 3′UTR. The 8 nt seed sequence is marked in yellow. B, C, Sequence (B) and secondary structure (C) of the COXIV 3′UTR, as determined by secondary structure prediction analysis (Mfold). The miR-338 target site (MTS) is indicated in yellow. D, The luciferase reporter plasmids carrying the firefly luciferase coding sequence (F-LUC) attached to the entire 3′UTR of rat COXIV (wild-type) or the 3′UTR in which the miR-338 target site was deleted (ΔMTS) are diagrammed. Luciferase expression was driven by the cytomegalovirus promoter (CMV). Luciferase activity of wild-type or ΔMTS COXIV 3′UTR reporter genes in the absence (control) or presence of the anti-miR-338 (25 n
m
), or a nontargeting (NT) anti-miR oligonucleotide (25 n
m
), is shown. Luciferase reporter constructs were cotransfected in equal ratios with the control plasmids coding for β-galactosidase (0.5 μg for each plasmid per transfection). The ratio of reporter to control plasmids in relative luminescence units was normalized for each reporter and plotted as a percentage of sham-transfected control value. Anti-miR oligonucleotide transfections were performed within 4 h following plasmid DNA transfection. Error bars represent the SEM for n = 10 cultures. *p < 0.05.
Figure 2.
Mature miR-338 is expressed in SCG neurons and is present in proximal and distal axons. A–D, Visualization of the presence of miR-338 at 21 d in culture by ISH in SCG axons (A), or soma and proximal axons (B); the scramble control probes show no significant staining in the soma or axons (C, D); scale bars, 10 μm. E, miR-338 levels increase during axonal outgrowth. The TaqMan miR Assay was used to quantify mature miR-338 isolated from either the soma and proximal axons (Soma), or distal axons (Axons) of SCG neurons at various DIV. Levels of miR-338 are expressed relative to β-actin mRNA which was used as an internal control. Error bars represent the SEM for n = 6 independent experiments. F, Samples obtained from the PCRs to quantify miR-338 in the distal axons (E) were size-fractionated on 4% agarose gels containing ethidium bromide and visualized using UV absorption (254 nm wavelength). Levels of β-actin mRNA served as an internal control. Negative control, PCR minus cDNA.
Figure 3.
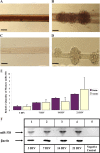
Localization of Dicer and eIF2c in the soma and axons of sympathetic neurons. A, Immunohistochemical examination of Dicer revealed granule-like structures visible in the soma and along the entire length of the axons of rat SCG neuronal cultures (15 DIV). B, Immunohistochemical analyses further visualized eIf2c in the axons and soma of SCG neurons using antibodies against this protein. Reactions conducted with Cy3-labeled secondary antibody alone produced only faint fluorescence. Scale bars: A, distal axons: 10 μm; soma and proximal axons: 50 μm; B, 50 μm.
Figure 4.
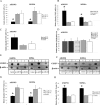
miRNA-338 reduces COXIV expression in SCG neurons. A, B, Quantification of COXIV mRNA levels in the soma and distal axons of SCG neurons transfected with 25 n
m
pre-miR-338 (A), or with 25 n
m
anti-miR-338 (B), as determined by qRT-PCR 24 h after oligonucleotide transfection. COXIV mRNA levels are expressed relative to β-actin mRNA. Error bars represent the SEM for n = 3 samples. *p < 0.05. qRT-PCR revealed a doubling in COXIV mRNA levels 4 h after anti-miR-338 transfection into the distal axons of SCG neurons (C). Transfection of SCG axons with precursor or anti-miR-338 oligonucleotides did not affect the relative abundance of COXII mRNA in the distal axons as shown by qRT-PCR using primers to amplify a 100 bp segment of COXII mRNA coding region (D). E, F, Immunoblots of axonal or somal protein lysates of SCG neurons transfected with miR-338 precursor (E), anti-miR-338 (F), or nontargeting short oligonucleotides (NT). β-Actin was used as a loading control. G, H, Immunoblots of axonal or somal protein lysates of SCG neurons transfected with either miR-338 precursor (G) or anti-miR-338 (H) were quantified using Image J. Quantification showed a significant change in COXIV protein levels under the influence of miR-338 (p < 0.05, t test in all comparisons).
Figure 5.
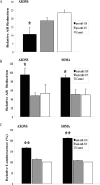
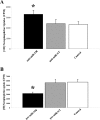
MiR-338 mediated alteration in COXIV levels modulates metabolic activity of mitochondria in the soma and distal axons of sympathetic neurons. A, B, SCG neurons were transfected with precursor miR-338 (A), anti-miR-338 (B), or nontargeting short oligonucleotides in the axon or soma compartments, respectively. AB (10%) was added to the culture media and cells were incubated for 24 h. AB data represent means ± SEM for three independent measures; *p < 0.05. ATP levels were measured in anti-miR-338 transfected axons or soma (C). Values were plotted in arbitrary luminescence units, using the luciferase cell viability assay. Data represent mean ± SEM; one-way ANOVA, **p < 0.0001.
Figure 6.
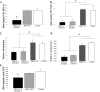
MiR-338 modulates norepinephrine uptake in the axons of sympathetic neurons. A, B, Distal axons were transfected with anti-miR-338 (A), precursor miR-338 (B), or nontargeting short oligonucleotides (NT). After transfection (24 h), axons were incubated for 120 min in the presence of [3H]NE (2 μCi/ml). NE uptake into Triton X-100 treated axons was measured by liquid scintillation spectrometry. *p < 0.0002. Norepinephrine uptake was subsequently assessed as outlined in A. Uptake data represent mean CPM ±SEM for six independent measures; *p < 0.002.
Figure 7.
SiRNA-mediated knockdown of axonal COXIV levels decreases axonal respiration and ATP levels and diminishes NE uptake in distal axons. Two independent siRNA oligonucleotides (25 n
m
) targeted against COXIV mRNA were introduced in distal axons of SCG neurons by lipofection and COXIV mRNA (A) and protein levels (B) quantitated 24 h later by qRT-PCR and immunoblot analyses, respectively. Knockdown of axonal COXIV expression reduced axonal oxygen consumption (C), ATP levels (D), and [3H]NE uptake (E). Values are expressed as mean ± SEM and statistical significance evaluated by one-way ANOVA. *p < 0.03.
Similar articles
- MicroRNA-338 regulates the axonal expression of multiple nuclear-encoded mitochondrial mRNAs encoding subunits of the oxidative phosphorylation machinery.
Aschrafi A, Kar AN, Natera-Naranjo O, MacGibeny MA, Gioio AE, Kaplan BB. Aschrafi A, et al. Cell Mol Life Sci. 2012 Dec;69(23):4017-27. doi: 10.1007/s00018-012-1064-8. Epub 2012 Jul 8. Cell Mol Life Sci. 2012. PMID: 22773120 Free PMC article. - Molecular determinants of cytochrome C oxidase IV mRNA axonal trafficking.
Kar AN, Vargas JNS, Chen CY, Kowalak JA, Gioio AE, Kaplan BB. Kar AN, et al. Mol Cell Neurosci. 2017 Apr;80:32-43. doi: 10.1016/j.mcn.2017.01.008. Epub 2017 Feb 1. Mol Cell Neurosci. 2017. PMID: 28161363 Free PMC article. - Regulation of axonal trafficking of cytochrome c oxidase IV mRNA.
Aschrafi A, Natera-Naranjo O, Gioio AE, Kaplan BB. Aschrafi A, et al. Mol Cell Neurosci. 2010 Apr;43(4):422-30. doi: 10.1016/j.mcn.2010.01.009. Epub 2010 Feb 6. Mol Cell Neurosci. 2010. PMID: 20144716 Free PMC article. - Axonal protein synthesis and the regulation of local mitochondrial function.
Kaplan BB, Gioio AE, Hillefors M, Aschrafi A. Kaplan BB, et al. Results Probl Cell Differ. 2009;48:225-42. doi: 10.1007/400_2009_1. Results Probl Cell Differ. 2009. PMID: 19343315 Free PMC article. Review. - Nuclear-Encoded Mitochondrial mRNAs: A Powerful Force in Axonal Growth and Development.
Gale JR, Aschrafi A, Gioio AE, Kaplan BB. Gale JR, et al. Neuroscientist. 2018 Apr;24(2):142-155. doi: 10.1177/1073858417714225. Epub 2017 Jun 14. Neuroscientist. 2018. PMID: 28614981 Review.
Cited by
- Identification of precursor microRNAs within distal axons of sensory neuron.
Kim HH, Kim P, Phay M, Yoo S. Kim HH, et al. J Neurochem. 2015 Jul;134(2):193-9. doi: 10.1111/jnc.13140. Epub 2015 May 23. J Neurochem. 2015. PMID: 25919946 Free PMC article. - Guidelines for mitochondrial RNA analysis.
Jusic A, Erpapazoglou Z, Dalgaard LT, Lakkisto P, de Gonzalo-Calvo D, Benczik B, Ágg B, Ferdinandy P, Fiedorowicz K, Schroen B, Lazou A, Devaux Y; EU-CardioRNA COST Action CA17129; AtheroNET COST Action CA21153. Jusic A, et al. Mol Ther Nucleic Acids. 2024 Jun 26;35(3):102262. doi: 10.1016/j.omtn.2024.102262. eCollection 2024 Sep 10. Mol Ther Nucleic Acids. 2024. PMID: 39091381 Free PMC article. Review. - MicroRNAs and deregulated gene expression networks in neurodegeneration.
Sonntag KC. Sonntag KC. Brain Res. 2010 Jun 18;1338:48-57. doi: 10.1016/j.brainres.2010.03.106. Epub 2010 Apr 7. Brain Res. 2010. PMID: 20380815 Free PMC article. Review. - Understanding neuronal connectivity through the post-transcriptional toolkit.
Loya CM, Van Vactor D, Fulga TA. Loya CM, et al. Genes Dev. 2010 Apr 1;24(7):625-35. doi: 10.1101/gad.1907710. Genes Dev. 2010. PMID: 20360381 Free PMC article. Review. - Dysregulation of the axonal trafficking of nuclear-encoded mitochondrial mRNA alters neuronal mitochondrial activity and mouse behavior.
Kar AN, Sun CY, Reichard K, Gervasi NM, Pickel J, Nakazawa K, Gioio AE, Kaplan BB. Kar AN, et al. Dev Neurobiol. 2014 Mar;74(3):333-50. doi: 10.1002/dneu.22141. Epub 2013 Nov 20. Dev Neurobiol. 2014. PMID: 24151253 Free PMC article.
References
- Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. - PubMed
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. - PubMed
- Ambros V, Chen X. The regulation of genes and genomes by small RNAs. Development. 2007;134:1635–1641. - PubMed
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials