Targeting the ubiquitin-proteasome system for cancer therapy - PubMed (original) (raw)
Review
Targeting the ubiquitin-proteasome system for cancer therapy
Yili Yang et al. Cancer Sci. 2009 Jan.
Abstract
The ubiquitin-proteasome system plays a critical role in controlling the level, activity and location of various cellular proteins. Significant progress has been made in investigating the molecular mechanisms of ubiquitination, particularly in understanding the structure of the ubiquitination machinery and identifying ubiquitin protein ligases, the primary specificity-determining enzymes. Therefore, it is now possible to target specific molecules involved in ubiquitination and proteasomal degradation to regulate many cellular processes such as signal transduction, proliferation and apoptosis. In particular, alterations in ubiquitination are observed in most, if not all, cancer cells. This is manifested by destabilization of tumor suppressors, such as p53, and overexpression of oncogenes such as c-Myc and c-Jun. In addition to the development and clinical validation of proteasome inhibitor, bortezomib, in myeloma therapy, recent studies have demonstrated that it is possible to develop inhibitors for specific ubiquitination and deubiquitination enzymes. With the help of structural studies, rational design and chemical synthesis, it is conceivable that we will be able to use 'druggable' inhibitors of the ubiquitin system to evaluate their effects in animal tumor models in the not-so-distant future.
Figures
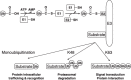
Figure 1
The ubiquitination cascade. Ubiquitin (Ub) is activated by E1 and conjugated to the active Cys of E1 through thioester bond. Activated Ub is then transferred to E2 that can bind with E3. An RING‐containing E3 facilitates the transfer of activated Ub to substrate from E2 directly, whereas a HECT domain E3 forms thioester bond with activated Ub and then transfer it to substrate. Monoubiquitination of target protein enable its recognition by many Ub‐recognizing domains in cells, leading to alteration of protein activity and location in cells. Formation of K48‐linked polyubiquitin chains on substrate proteins result in their degradation in proteasomes. Formation of K63‐linked polyubiquitin chains are involved in cellular processes such as signal transduction and DNA repair. AMP, adenosine triphosphate; ATP, adenosine monophosphate.
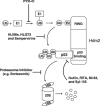
Figure 2
Inhibitors that block the Hdm2‐mediated ubiquitination and proteasomal degradation of p53.
Similar articles
- Ubiquitin and cancer: from molecular targets and mechanisms to the clinic -- AACR Special Conference.
Colland F. Colland F. IDrugs. 2006 Mar;9(3):179-81. IDrugs. 2006. PMID: 16523381 No abstract available. - Targeting the ubiquitin-proteasome system to activate wild-type p53 for cancer therapy.
Allende-Vega N, Saville MK. Allende-Vega N, et al. Semin Cancer Biol. 2010 Feb;20(1):29-39. doi: 10.1016/j.semcancer.2009.10.004. Epub 2009 Nov 6. Semin Cancer Biol. 2010. PMID: 19897040 Review. - Ubiquitination and deubiquitination: Implications on cancer therapy.
Dagar G, Kumar R, Yadav KK, Singh M, Pandita TK. Dagar G, et al. Biochim Biophys Acta Gene Regul Mech. 2023 Dec;1866(4):194979. doi: 10.1016/j.bbagrm.2023.194979. Epub 2023 Aug 24. Biochim Biophys Acta Gene Regul Mech. 2023. PMID: 37633647 Review. - Chemical approaches to targeted protein degradation through modulation of the ubiquitin-proteasome pathway.
Collins I, Wang H, Caldwell JJ, Chopra R. Collins I, et al. Biochem J. 2017 Mar 15;474(7):1127-1147. doi: 10.1042/BCJ20160762. Biochem J. 2017. PMID: 28298557 Free PMC article. Review. - The therapeutic potential of deubiquitinating enzyme inhibitors.
Colland F. Colland F. Biochem Soc Trans. 2010 Feb;38(Pt 1):137-43. doi: 10.1042/BST0380137. Biochem Soc Trans. 2010. PMID: 20074048 Review.
Cited by
- Functional proteomics to dissect tyrosine kinase signalling pathways in cancer.
Kolch W, Pitt A. Kolch W, et al. Nat Rev Cancer. 2010 Sep;10(9):618-29. doi: 10.1038/nrc2900. Epub 2010 Aug 19. Nat Rev Cancer. 2010. PMID: 20720570 Review. - Overview of Targeted Drugs for Mature B-Cell Non-hodgkin Lymphomas.
Crisci S, Di Francia R, Mele S, Vitale P, Ronga G, De Filippi R, Berretta M, Rossi P, Pinto A. Crisci S, et al. Front Oncol. 2019 Jun 4;9:443. doi: 10.3389/fonc.2019.00443. eCollection 2019. Front Oncol. 2019. PMID: 31214498 Free PMC article. Review. - Insights Into the Properties, Biological Functions, and Regulation of USP21.
An T, Lu Y, Yan X, Hou J. An T, et al. Front Pharmacol. 2022 Jun 30;13:944089. doi: 10.3389/fphar.2022.944089. eCollection 2022. Front Pharmacol. 2022. PMID: 35846989 Free PMC article. Review. - Negative regulation of DAB2IP by Akt and SCFFbw7 pathways.
Dai X, North BJ, Inuzuka H. Dai X, et al. Oncotarget. 2014 May 30;5(10):3307-15. doi: 10.18632/oncotarget.1939. Oncotarget. 2014. PMID: 24912918 Free PMC article. - Targeting NF-κB in infantile hemangioma-derived stem cells reduces VEGF-A expression.
Greenberger S, Adini I, Boscolo E, Mulliken JB, Bischoff J. Greenberger S, et al. Angiogenesis. 2010 Dec;13(4):327-35. doi: 10.1007/s10456-010-9189-6. Epub 2010 Sep 25. Angiogenesis. 2010. PMID: 20872175 Free PMC article.
References
- Hershko A. Ubiquitin. roles in protein modification and breakdown. Cell 1983; 34: 11–12. - PubMed
- Cadwell K, Coscoy L. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science 2005; 309: 127–30. - PubMed
- Hoppe T. Multiubiquitylation by E4 enzymes: ‘one size’ doesn't fit all. Trends Biochem Sci 2005; 30: 183–7. - PubMed
- Hoeller D, Hecker CM, Wagner S, Rogov V, Dotsch V, Dikic I. E3‐independent monoubiquitination of ubiquitin‐binding proteins. Mol Cell 2007; 26: 891–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous