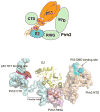Molecular basis of Pirh2-mediated p53 ubiquitylation - PubMed (original) (raw)
. 2008 Dec;15(12):1334-42.
doi: 10.1038/nsmb.1521. Epub 2008 Nov 30.
Rob C Laister, Alexander Lemak, Bin Wu, Elizabeth Tai, Shili Duan, Jonathan Lukin, Maria Sunnerhagen, Sampath Srisailam, Murthy Karra, Sam Benchimol, Cheryl H Arrowsmith
Affiliations
- PMID: 19043414
- PMCID: PMC4075976
- DOI: 10.1038/nsmb.1521
Molecular basis of Pirh2-mediated p53 ubiquitylation
Yi Sheng et al. Nat Struct Mol Biol. 2008 Dec.
Abstract
Pirh2 (p53-induced RING-H2 domain protein; also known as Rchy1) is an E3 ubiquitin ligase involved in a negative-feedback loop with p53. Using NMR spectroscopy, we show that Pirh2 is a unique cysteine-rich protein comprising three modular domains. The protein binds nine zinc ions using a variety of zinc coordination schemes, including a RING domain and a left-handed beta-spiral in which three zinc ions align three consecutive small beta-sheets in an interleaved fashion. We show that Pirh2-p53 interaction is dependent on the C-terminal zinc binding module of Pirh2, which binds to the tetramerization domain of p53. As a result, Pirh2 preferentially ubiquitylates the tetrameric form of p53 in vitro and in vivo, suggesting that Pirh2 regulates protein turnover of the transcriptionally active form of p53.
Figures
Figure 1
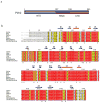
The sequence and domain organization of Pirh2. (a) Schematic diagram illustrating the boundaries for the three domains of Pirh2 with the NTD in gray, the RING in blue and the CTD in brown. (b) The multiple sequence alignment of Pirh2 from eukaryotic organisms with the secondary structure elements of human Pirh2 are indicated above the sequence. Arrows designate β strands, solenoids for α-helices and asterisks indicating zinc coordinating cysteins and histidines. The secondary structure elements are colored in gray, blue and brown, representing NTD, RING and CTD, respectively. The alignment was generated by CLUSTAL W and was printed using the ESPript 2.2 software package.
Figure 2
Ribbon representations of the three individual Pirh2 domains and the zinc coordination schemes of Pirh2 (a) the Pirh2 NTD. The N lobe of the Pirh2 NTD showing the interleaved Zn1 and Zn2 sites coordinated by a sequence motif CHC4H2 and the Zn3 site coordinated by four cysteines. The C lobe of the Pirh2 NTD showing the left-handed β spiral structure with the three CCHC motifs coordinating Zn4, Zn5 and Zn6. (b) The Pirh2 RING domain showing the interleaved Zn7 and Zn8 sites coordinated by the sequence motif C3H2C3. (c) The Pirh2 CTD showing the Zn9 site coordinated by four cysteines. The nine zinc atoms are shown as spheres.
Figure 3
Pirh2 interacts with p53 via a two-site binding mode. (a) GST pull-down assays of GST-Pirh2 fusion proteins with the full-length p53. (b) GST pull-down assays of GST-Pirh2 NTD and GST-Pirh2 CTD fusions with purified p53 domains as indicated. The labeled lanes reflect loaded material (L), column flow-through after wash (W) and elutate (E). (c) Surface representations of the Pirh2 NTD (c) and CTD (d) showing the p53 binding interface, as determined by NMR chemical shift perturbation experiments. The residues whose resonances show substantial shifts upon addition of unlabeled p53 into 15N labeled Pirh2 samples are indicated (see Supplementary Fig. 3 and 4 online).
Figure 4
Mapping UBE2D2/UbcH5b binding interface. (a) Surface representation of the Pirh2 RING domain showing the UBE2D2/UbcH5b binding interface, as determined by NMR chemical shift perturbation experiments. Pro146 and Pro184, which lack amide resonances, are also likely to be involved (see Supplementary Fig. 2 online). (b) Composite chemical shift changes versus residue number for Pirh2 RING domain upon binding to UBE2D2/UbcH5b. The values shown were calculated by using the equation Δcomp = [ΔδHN2 + (ΔδN/5)2]1/2. The inset shows the chemical shift changes of the representative residues upon titration with UBE2D2/UbcH5b.
Figure 5
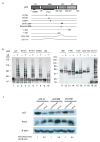
Pirh2-mediated ubiquitilyation of both itself and p53. (a) Schematic diagram of the p53 mutants used in panel c. (b) Pirh2 mediated in vitro ubiquitylation assays of both wild type and mutant p53 constructs in the presence or absence of ubiquitin (UB). The reaction products were resolved on a 7.5% SDS-PAGE gel for lanes 1–10 and a 10% gel for lanes 11–20. Detection of p53 ubquitylation was performed as described in Methods. (c) Pirh2 expression results in reduced levels of tetrameric p53 in vivo. H1299 cells were transfected with either full length hPIRH2 or an empty vector control (pcDNA3(neo)), along with either tetrameric (p53 WT), dimeric (p53 DM) or monomeric (p53 MM) p53. Equal amounts of cell lysate were analyzed by SDS-PAGE and the amounts of p53, Pirh2 and β-actin were detected by immunoblot. The p53 (WT, DM and MM) levels were quantified as the ratio of p53 intensity to β-actin intensity. For each empty vector control experiment the p53 level was normalized to 1 in order to reflect the background level of endogenous Pirh2 activity against each type of transfected p53. The p53 levels (WT, DM and MM) in Pirh2 co-transfected cells are intensity ratio relative to the appropriate control experiment.
Figure 6
Assessment of the ubiquitylation activity of Pirh2 deletion mutants. (a) Schematic drawing of the Pirh2 deletion mutants and their activities in auto- and p53-ubiquitylation. (b) Autoubiquitylation (lanes 1–10, left panel) and ubiquitylation of p53 (lanes 11–20, right panel) using different Pirh2 deletion mutants as indicated. The degradation product of the GST-Pirh2 fusions to GST alone is noted (GST). (c) Ubiquitylation of p53 using various concentrations of the wild type Pirh2, the mutants 138–261 and 1–195 as indicated. (d) Ubiquitylation of p53 using the wild type Pirh2, and the mutants 138–261, 138–253, C186A and M176E. All Pirh2 mutants were purified as GST fusions and subjected to the in vitro ubiquitylation assays in the absence (lanes 1–10) and the presence of p53 (lanes 11–20). Autoubiquitylation of the Pirh2 mutants was evaluated by blotting with an antibody against GST. Ubiquitylation of p53 mediated by the Pirh2 mutants was evaluated by blotting with an antibody against p53 (Pab1801).
Figure 7
A model of a potential ternary complex between Pirh2, p53 and the E2 enzyme UBE2D2/UbcH5b. The top panel illustrates schematically how domains of tetrameric p53 interact with those of Pirh2. The UBE2D2-Pirh2 ring domain orientations are based on the crystal structure of the homologous complex formed between UBCH7 and the c-Cbl ring domain. The active site cysteine of E2D2 which is charged with ubiquitin is shown in red.
Similar articles
- Identification and characterization of two novel isoforms of Pirh2 ubiquitin ligase that negatively regulate p53 independent of RING finger domains.
Corcoran CA, Montalbano J, Sun H, He Q, Huang Y, Sheikh MS. Corcoran CA, et al. J Biol Chem. 2009 Aug 14;284(33):21955-21970. doi: 10.1074/jbc.M109.024232. Epub 2009 May 29. J Biol Chem. 2009. PMID: 19483087 Free PMC article. - Structural and functional comparison of the RING domains of two p53 E3 ligases, Mdm2 and Pirh2.
Shloush J, Vlassov JE, Engson I, Duan S, Saridakis V, Dhe-Paganon S, Raught B, Sheng Y, Arrowsmith CH. Shloush J, et al. J Biol Chem. 2011 Feb 11;286(6):4796-808. doi: 10.1074/jbc.M110.157669. Epub 2010 Nov 17. J Biol Chem. 2011. PMID: 21084285 Free PMC article. - Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation.
Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, Parant JM, Lozano G, Hakem R, Benchimol S. Leng RP, et al. Cell. 2003 Mar 21;112(6):779-91. doi: 10.1016/s0092-8674(03)00193-4. Cell. 2003. PMID: 12654245 - The Role of E3 Ligase Pirh2 in Disease.
Daks A, Fedorova O, Parfenyev S, Nevzorov I, Shuvalov O, Barlev NA. Daks A, et al. Cells. 2022 Apr 30;11(9):1515. doi: 10.3390/cells11091515. Cells. 2022. PMID: 35563824 Free PMC article. Review. - Pirh2 RING-finger E3 ubiquitin ligase: its role in tumorigenesis and cancer therapy.
Jung YS, Qian Y, Chen X. Jung YS, et al. FEBS Lett. 2012 May 21;586(10):1397-402. doi: 10.1016/j.febslet.2012.03.052. Epub 2012 Apr 3. FEBS Lett. 2012. PMID: 22673504 Free PMC article. Review.
Cited by
- TMEM209 promotes hepatocellular carcinoma progression by activating the Wnt/β-catenin signaling pathway through KPNB1 stabilization.
Fang H, Shi X, Gao J, Yan Z, Wang Y, Chen Y, Zhang J, Guo W. Fang H, et al. Cell Death Discov. 2024 Oct 16;10(1):438. doi: 10.1038/s41420-024-02207-9. Cell Death Discov. 2024. PMID: 39414762 Free PMC article. - RCHY1 and OPTN are required for melanophagy, selective autophagy of melanosomes.
Lee KW, Ryu KJ, Kim M, Lim S, Kim J, Kim JY, Hwangbo C, Yoo J, Cho YY, Kim KD. Lee KW, et al. Proc Natl Acad Sci U S A. 2024 Apr 2;121(14):e2318039121. doi: 10.1073/pnas.2318039121. Epub 2024 Mar 27. Proc Natl Acad Sci U S A. 2024. PMID: 38536750 Free PMC article. - Mechanism and evolutionary origins of alanine-tail C-degron recognition by E3 ligases Pirh2 and CRL2-KLHDC10.
Patil PR, Burroughs AM, Misra M, Cerullo F, Costas-Insua C, Hung HC, Dikic I, Aravind L, Joazeiro CAP. Patil PR, et al. Cell Rep. 2023 Sep 26;42(9):113100. doi: 10.1016/j.celrep.2023.113100. Epub 2023 Sep 6. Cell Rep. 2023. PMID: 37676773 Free PMC article. - Zooming into the structure-function of RING finger proteins for anti-cancer therapeutic applications.
George M, Masamba P, Iwalokun BA, Kappo AP. George M, et al. Am J Cancer Res. 2023 Jul 15;13(7):2773-2789. eCollection 2023. Am J Cancer Res. 2023. PMID: 37559981 Free PMC article. Review. - The ubiquitin-protein ligase MIEL1 localizes to peroxisomes to promote seedling oleosin degradation and lipid droplet mobilization.
Traver MS, Bartel B. Traver MS, et al. Proc Natl Acad Sci U S A. 2023 Jul 18;120(29):e2304870120. doi: 10.1073/pnas.2304870120. Epub 2023 Jul 6. Proc Natl Acad Sci U S A. 2023. PMID: 37410814 Free PMC article.
References
- Vousden KH, Prives C. P53 and prognosis: new insights and further complexity. Cell. 2005;120:7–10. - PubMed
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–31. - PubMed
- Michael D, Oren M. The p53-Mdm2 module and the ubiquitin system. Semin Cancer Biol. 2003;13:49–58. - PubMed
- Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–7. - PubMed
- Chene P. Inhibiting the p53-MDM2 interaction: an important target for cancer therapy. Nat Rev Cancer. 2003;3:102–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50-GM62413-01/GM/NIGMS NIH HHS/United States
- P50 GM062413-01/GM/NIGMS NIH HHS/United States
- U54 GM074958-05/GM/NIGMS NIH HHS/United States
- P50 GM062413/GM/NIGMS NIH HHS/United States
- U54 GM074958/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous