snoRNA, a novel precursor of microRNA in Giardia lamblia - PubMed (original) (raw)
snoRNA, a novel precursor of microRNA in Giardia lamblia
Ashesh A Saraiya et al. PLoS Pathog. 2008 Nov.
Abstract
An Argonaute homolog and a functional Dicer have been identified in the ancient eukaryote Giardia lamblia, which apparently lacks the ability to perform RNA interference (RNAi). The Giardia Argonaute plays an essential role in growth and is capable of binding specifically to the m(7)G-cap, suggesting a potential involvement in microRNA (miRNA)-mediated translational repression. To test such a possibility, small RNAs were isolated from Giardia trophozoites, cloned, and sequenced. A 26-nucleotide (nt) small RNA (miR2) was identified as a product of Dicer-processed snoRNA GlsR17 and localized to the cytoplasm by fluorescence in situ hybridization, whereas GlsR17 was found primarily in the nucleolus of only one of the two nuclei in Giardia. Three other small RNAs were also identified as products of snoRNAs, suggesting that the latter could be novel precursors of miRNAs in Giardia. Putative miR2 target sites were identified at the 3'-untranslated regions (UTR) of 22 variant surface protein mRNAs using the miRanda program. In vivo expression of Renilla luciferase mRNA containing six identical miR2 target sites in the 3'-UTR was reduced by 40% when co-transfected with synthetic miR2, while the level of luciferase mRNA remained unaffected. Thus, miR2 likely affects translation but not mRNA stability. This repression, however, was not observed when Argonaute was knocked down in Giardia using a ribozyme-antisense RNA. Instead, an enhancement of luciferase expression was observed, suggesting a loss of endogenous miR2-mediated repression when this protein is depleted. Additionally, the level of miR2 was significantly reduced when Dicer was knocked down. In all, the evidence indicates the presence of a snoRNA-derived miRNA-mediated translational repression in Giardia.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Figure 1. MFOLD analysis of snoRNAs GlsR16 and GlsR17.
The sequences of mature snoRNA GlsR16 and the 64 nt 3′-portion of GlsR17 were submitted to the MFOLD RNA server. Both snoRNAs produced secondary structures that are suitable for being Dicer substrates. The red letters represent miR3 in GlsR16 and miR2 in GlsR17. The Box C (blue) and Box D (green) motifs are indicated. Both miRNAs are located at the 3′-end of their precursors.
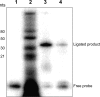
Figure 2. Splinted ligation of miR2.
Size fractionated RNA (<40 nts) was analyzed by splinted ligation for the presence of miR2. (Lane 1) Negative control containing only the labeled RNA probe. (Lane 2) RNA markers. (Lane 3) Positive control using a synthetic miR2. (Lane 4) Size fractionated RNA (200 ng) from Giardia WB trophozoites. The presence of a discrete band migrating to the same position with the positive control suggests the presence of miR2 in the small RNA population.
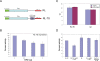
Figure 3. miR2-mediated translational repression of mRNA with putative target site at the 3′-UTR.
(A) Diagram of the reporter constructs RL and RL-TS. (B) Percent luciferase activity of Giardia trophozoites transfected with RL-TS plus different concentrations of exogenous miR2. These results represent the average number and standard deviation of at least two replicates. (C) Real time quantitative RT-PCR comparison of RL-TS mRNA concentrations in the presence and absence of exogenously introduced miR2. Ran mRNA was monitored as a control. The data represent the average number and standard deviation from three independent transfection experiments. (D) Percent luciferase activity in RL-TS and RL transfected cells. The introduction of miR2 (1.0 µg) represses RL-TS activity by 35%. The absence of target sites (TS) in RL results in a small increase of expression by ∼15% that is not repressed by co-transfection with miR2. This suggests that the target sites are required for repression of luciferase activity by both endogenous and exogenous miR2. The use of a mutant miR2, miR2neg, does not affect the luciferase activity, indicating a specificity between miR2 and its target sites. These data represent the average number and standard deviation of four independent transfection experiments.
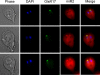
Figure 4. Localization of GlsR17 and miR2 in Giardia using FISH.
GlsR17 is predominately localized to a single nucleus with a focus in the putative nucleolus in each Giardia trophozoite. miR2 is localized primarily to the cytoplasm.
Figure 5. Effects of Dicer knockdown on levels of miR2 and miR3 in Giardia monitored by splinted ligation.
(A) Lane 1, pegative control. Lane 2, positive control using a synthetic sample of miR2. Lane 3, a 33 ng sample of size fractionated small RNA (<40 nts) from giardiavirus-infected Giardia WB trophozoites transfected with an empty vector. Lane 4, a 33 ng sample of size fractionated small RNA (<40 nts) from giardiavirus-infected Giardia WB trophozoites transfected with the Dicer ribozyme. (B) Lane 1, positive control using a sample of synthetic miR3. Lane 2, a 33 ng sample of size fractionated small RNA (<40 nts) from giardiavirus-infected Giardia WB trophozoites transfected with an empty vector. Lane 3, a 33 ng sample of size fractionated small RNA (<40 nts) from giardiavirus-infected Giardia WB trophozoites transfected with the Dicer ribozyme. (C) Denatured PAGE analysis of size-fractionated RNA (<200 nts) stained with SYBR Gold. Lane 1, control cells; lane 2, dicer knockdown cells.
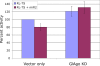
Figure 6. Loss of miR2 repression in GlAgo knockdown Giardia trophozoites.
The translation of RL-TS in Giardia cells is repressed by introducing exogenous miR2 (1 µg) into the cells but not in the cells where GlAgo is partially knocked down. KD, knockdown.
Similar articles
- Gene regulation in Giardia lambia involves a putative microRNA derived from a small nucleolar RNA.
Li W, Saraiya AA, Wang CC. Li W, et al. PLoS Negl Trop Dis. 2011 Oct;5(10):e1338. doi: 10.1371/journal.pntd.0001338. Epub 2011 Oct 18. PLoS Negl Trop Dis. 2011. PMID: 22028939 Free PMC article. - A microRNA derived from an apparent canonical biogenesis pathway regulates variant surface protein gene expression in Giardia lamblia.
Saraiya AA, Li W, Wang CC. Saraiya AA, et al. RNA. 2011 Dec;17(12):2152-64. doi: 10.1261/rna.028118.111. Epub 2011 Oct 27. RNA. 2011. PMID: 22033329 Free PMC article. - The profile of snoRNA-derived microRNAs that regulate expression of variant surface proteins in Giardia lamblia.
Li W, Saraiya AA, Wang CC. Li W, et al. Cell Microbiol. 2012 Sep;14(9):1455-73. doi: 10.1111/j.1462-5822.2012.01811.x. Epub 2012 May 23. Cell Microbiol. 2012. PMID: 22568619 Free PMC article. - Exploration of Giardia small nucleolar RNAs (snoRNAs) and their possible microRNA derivatives.
Lagunas-Rangel FA. Lagunas-Rangel FA. Parasitology. 2024 May;151(6):539-545. doi: 10.1017/S003118202400060X. Epub 2024 May 20. Parasitology. 2024. PMID: 38767317 Free PMC article. Review. - The many faces of small nucleolar RNAs.
Bratkovič T, Rogelj B. Bratkovič T, et al. Biochim Biophys Acta. 2014 Jun;1839(6):438-43. doi: 10.1016/j.bbagrm.2014.04.009. Epub 2014 Apr 13. Biochim Biophys Acta. 2014. PMID: 24735946 Review.
Cited by
- Global profiling of miRNAs and the hairpin precursors: insights into miRNA processing and novel miRNA discovery.
Li N, You X, Chen T, Mackowiak SD, Friedländer MR, Weigt M, Du H, Gogol-Döring A, Chang Z, Dieterich C, Hu Y, Chen W. Li N, et al. Nucleic Acids Res. 2013 Apr 1;41(6):3619-34. doi: 10.1093/nar/gkt072. Epub 2013 Feb 8. Nucleic Acids Res. 2013. PMID: 23396444 Free PMC article. - Regulation of microRNA biogenesis and turnover by animals and their viruses.
Libri V, Miesen P, van Rij RP, Buck AH. Libri V, et al. Cell Mol Life Sci. 2013 Oct;70(19):3525-44. doi: 10.1007/s00018-012-1257-1. Epub 2013 Jan 26. Cell Mol Life Sci. 2013. PMID: 23354060 Free PMC article. Review. - Comparative transcriptome analysis of small noncoding RNAs in different stages of Trypanosoma brucei.
Zheng LL, Wen YZ, Yang JH, Liao JY, Shao P, Xu H, Zhou H, Wen JZ, Lun ZR, Ayala FJ, Qu LH. Zheng LL, et al. RNA. 2013 Jul;19(7):863-75. doi: 10.1261/rna.035683.112. Epub 2013 May 23. RNA. 2013. PMID: 23704326 Free PMC article. - microRNAs in parasites and parasite infection.
Zheng Y, Cai X, Bradley JE. Zheng Y, et al. RNA Biol. 2013 Mar;10(3):371-9. doi: 10.4161/rna.23716. Epub 2013 Feb 7. RNA Biol. 2013. PMID: 23392243 Free PMC article. Review. - Live imaging of mitosomes and hydrogenosomes by HaloTag technology.
Martincová E, Voleman L, Najdrová V, De Napoli M, Eshar S, Gualdron M, Hopp CS, Sanin DE, Tembo DL, Van Tyne D, Walker D, Marcinčiková M, Tachezy J, Doležal P. Martincová E, et al. PLoS One. 2012;7(4):e36314. doi: 10.1371/journal.pone.0036314. Epub 2012 Apr 27. PLoS One. 2012. PMID: 22558433 Free PMC article.
References
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. - PubMed
- Pasquinelli AE, Ruvkun G. Control of developmental timing by micrornas and their targets. Annu Rev Cell Dev Biol. 2002;18:495–513. - PubMed
- Miska EA. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev. 2005;15:563–568. - PubMed
- Lee Y, Ahn C, Han J, Choi H, Kim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous