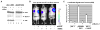The ARF tumor suppressor can promote the progression of some tumors - PubMed (original) (raw)
The ARF tumor suppressor can promote the progression of some tumors
Olivier Humbey et al. Cancer Res. 2008.
Abstract
p14/p19ARF (ARF) is a tumor suppressor gene that is frequently mutated in human cancer. ARF has multiple tumor suppressor functions, some of which are mediated by signaling to p53. Surprisingly, a significant fraction of human tumors retain persistently high levels of ARF, suggesting that ARF may possess a prosurvival function. We show that ARF protein is markedly up-regulated in cells exposed to nutrient starvation. Cells with silenced ARF show reduced autophagy and reduced viability when placed under conditions of starvation. We show for the first time that ARF silencing can limit the progression of some tumors, such as lymphoma, but not others, such as E1A/Ras-induced tumors. Specifically, myc-driven lymphomas with mutant p53 tend to overexpress ARF; we show that silencing ARF in these tumors greatly impedes their progression. These data are the first to show that ARF can act in a p53-independent manner to promote the progression of some tumors.
Figures
Figure 1. Nutrient deprivation causes significant upregulation of ARF, along with mitochondrial localization of ARF
(A) Western analysis of whole cell lysate from p53 −/− MEFs (passage 3) untreated or incubated in Hank’s Buffered Salt Solution (HBSS) for the indicated timepoints (starvation) using antisera to actin (loading control) and ARF. (B) Western analysis of MEFs with wt p53 (passage 2), untreated or incubated in Hank’s Buffered Salt Solution (HBSS) for the indicated timepoints (starvation) using antisera to actin (loading control) and ARF. (C) Analysis of the half life of ARF in untreated and nutrient-deprived cells. p53-null MEFs were untreated (UNT) or incubated for 2 h in HBSS, followed by supplementation with complete media containing 40 ug/uL cycloheximide to halt new protein synthesis. Cells were harvested and analyzed by western blotting for ARF at the indicated timepoints; the data depicted are representative of two independent experiments. (D) Immuno-electron microscopy using ARF antisera followed by Protein G-Gold in mouse embryo fibroblasts from the p53-knockout mouse following two hours treatment with HBSS (starvation). Arrows point to gold particles in mitochondria and nucleoli. Scale bar is 500 nm.
Figure 2. ARF-silencing reduces starvation-induced autophagy and survival
(A) Western analysis for LC3 II in p53 −/− MEFs following nutrient starvation (HBSS). Cells were infected for 48 hours with a short hairpin for p53 (shp53, negative control) or short hairpin for ARF (shARF), and incubated for 2 hrs in HBSS. Data depicted are representative of three independent experiments; the depicted data were obtained from a single blot that was cropped for optimal data presentation. The difference in LC3 II in unstressed cells in lanes 1 and 3 was not consistent or reproducible from experiment to experiment. (B) Electron microscopy of p53-null MEFs infected with shp53 (negative control, top panels) or shARF (bottom panels) following 2 hour incubation in HBSS (starvation). Scale bars: Left panels, 5 um. Right panels 500 nm. (C) The degradation of long-lived proteins in p53-null MEFs infected with short hairpin to p53 (negative control, black line) is increased compared to those infected with short hairpin to ARF (shARF, gray line) following starvation in HBSS for 24 hours; results are normalized to cell number. The x axis indicates timepoints of sampling. Data are representative of two independent experiments. (D) The viability of p53-null cells infected with short hairpin to p53 (MEF p53 −/−) or shARF following 24 hour treatment with HBSS is depicted; equal numbers of viable cells were plated at the 0 timepoint (1 × 106), and cell viability was determined using the ViaCount assay and the Guava Personal Cell Analysis machine. Data depicted are the averaged results from three independent experiments plus standard error.
Figure 3. Tumors without ARF are more sensitive to metabolic stress (silencing of Beclin-1)
(A) Western analysis of p53-null MEFS (high ARF) and MEFs from the p53/ARF double knockout mouse (p53/ARF DKO) following infection with short hairpin to silence Beclin-1 (shBec1) for the levels of actin (loading control), ARF and Beclin-1. (B) Bioluminescent imaging of xenograft tumors derived from early passage MEFs from the p53 −/− mouse, or p53/ARF double knockout (DKO) stably-infected with luciferase virus, and then infected with the retroviruses indicated (shARF or shBec) for 48 hours. Tumors were imaged after 21 days; equivalent results were seen after 30 days, and all measurements were consistent with caliper measurements. (C) Analysis of the combined imaging data from two independent experiments depicted in B, using three mice for each sample per experiment (6 samples total per retrovirus combination); all values represent the raw mean value of bioluminescence (mean BLI) along with standard error of the mean (SEM).
Figure 4. Silencing of ARF inhibits lymphoma development
(A) Bioluminescent imaging of tail-vein injected, luciferase-expressing B2998 B-cell lymphoma cells from the Eμ-myc mouse, infected with vector control or two different short hairpins for ARF, shARF157 or shARF56. Most tumor cells track to the lymph nodes and meninges. Results are at day 7 following tail vein injection; equivalent results were obtained at day 14. Data depicted are representative of two independent experiments, using 3 mice for each retrovirus (shControl, shARF157, shARF56). The middle panel depicts western analysis for p16, ARF and actin control in infected lymphoma cells from the B2998 lymphoma, 48 hours after retroviral infection. The right panel depicts the combined data from 2 independent experiments using 3 mice per virus, along with standard error of the mean. Silencing of ARF, but not infection with short hairpin control vector, reduces tumor volume by 60% (p<0.01). (B) The viability of B2998 cells with silenced ARF is decreased following HBSS treatment for 24 hours. Cell viability was determined using the ViaCount assay and the Guava Personal Cell Analysis machine. Data depicted are the averaged results from three independent experiments plus standard error. The p value reflects comparison between the second and fourth column. (C) Silencing ARF in B2998 lymphoma cells reduces the steady state level of autophagy. Western analysis for ARF, actin and p62SQSTM1 (which is degraded by autophagy) in cells infected with shControl or each of two ARF short hairpins (shARF157 and shARF56). (D) Silencing ARF in a primary T cell lymphoma from the p53 −/− mouse impedes tumor progression. Results are at day 2 following tail vein injection; equivalent results were obtained at day 4. Results are representative of two independent experiments. The right panel depicts western analysis for ARF and loading control (actin).
Similar articles
- Regulation of the Arf tumor suppressor in Emicro-Myc transgenic mice: longitudinal study of Myc-induced lymphomagenesis.
Bertwistle D, Sherr CJ. Bertwistle D, et al. Blood. 2007 Jan 15;109(2):792-4. doi: 10.1182/blood-2006-07-033985. Epub 2006 Sep 12. Blood. 2007. PMID: 16968893 - The inhibitor of cyclin-dependent kinase 4a/alternative reading frame (INK4a/ARF) locus encoded proteins p16INK4a and p19ARF repress cyclin D1 transcription through distinct cis elements.
D'Amico M, Wu K, Fu M, Rao M, Albanese C, Russell RG, Lian H, Bregman D, White MA, Pestell RG. D'Amico M, et al. Cancer Res. 2004 Jun 15;64(12):4122-30. doi: 10.1158/0008-5472.CAN-03-2519. Cancer Res. 2004. PMID: 15205322 - Frequent alterations of the p14(ARF) and p16(INK4a) genes in primary central nervous system lymphomas.
Nakamura M, Sakaki T, Hashimoto H, Nakase H, Ishida E, Shimada K, Konishi N. Nakamura M, et al. Cancer Res. 2001 Sep 1;61(17):6335-9. Cancer Res. 2001. PMID: 11522621 - p53-Dependent and -independent functions of the Arf tumor suppressor.
Sherr CJ, Bertwistle D, DEN Besten W, Kuo ML, Sugimoto M, Tago K, Williams RT, Zindy F, Roussel MF. Sherr CJ, et al. Cold Spring Harb Symp Quant Biol. 2005;70:129-37. doi: 10.1101/sqb.2005.70.004. Cold Spring Harb Symp Quant Biol. 2005. PMID: 16869746 Review. - Genetic dissection of melanoma pathways in the mouse.
Yang FC, Merlino G, Chin L. Yang FC, et al. Semin Cancer Biol. 2001 Jun;11(3):261-8. doi: 10.1006/scbi.2000.0376. Semin Cancer Biol. 2001. PMID: 11407950 Review.
Cited by
- Mutation of the Conserved Threonine 8 within the Human ARF Tumour Suppressor Protein Regulates Autophagy.
Fontana R, Guidone D, Angrisano T, Calabrò V, Pollice A, La Mantia G, Vivo M. Fontana R, et al. Biomolecules. 2022 Jan 13;12(1):126. doi: 10.3390/biom12010126. Biomolecules. 2022. PMID: 35053274 Free PMC article. - A conserved domain in exon 2 coding for the human and murine ARF tumor suppressor protein is required for autophagy induction.
Budina-Kolomets A, Hontz RD, Pimkina J, Murphy ME. Budina-Kolomets A, et al. Autophagy. 2013 Oct;9(10):1553-65. doi: 10.4161/auto.25831. Epub 2013 Aug 7. Autophagy. 2013. PMID: 23939042 Free PMC article. - Power of PTEN/AKT: Molecular switch between tumor suppressors and oncogenes.
Xie Y, Naizabekov S, Chen Z, Tokay T. Xie Y, et al. Oncol Lett. 2016 Jul;12(1):375-378. doi: 10.3892/ol.2016.4636. Epub 2016 May 26. Oncol Lett. 2016. PMID: 27347153 Free PMC article. - Resistance and gain-of-resistance phenotypes in cancers harboring wild-type p53.
Martinez-Rivera M, Siddik ZH. Martinez-Rivera M, et al. Biochem Pharmacol. 2012 Apr 15;83(8):1049-62. doi: 10.1016/j.bcp.2011.12.026. Epub 2011 Dec 26. Biochem Pharmacol. 2012. PMID: 22227014 Free PMC article. Review. - PKC Dependent p14ARF Phosphorylation on Threonine 8 Drives Cell Proliferation.
Fontana R, Guidone D, Sangermano F, Calabrò V, Pollice A, La Mantia G, Vivo M. Fontana R, et al. Sci Rep. 2018 May 4;8(1):7056. doi: 10.1038/s41598-018-25496-4. Sci Rep. 2018. PMID: 29728595 Free PMC article.
References
- Sherr CJ, Bertwistle D, Den Besten W, et al. p53-Dependent and -independent functions of the Arf tumor suppressor. Cold Spring Harb Symp Quant Biol. 2005;70:129–37. - PubMed
- Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. - PubMed
- Reef S, Zalckvar E, Shifman O, et al. A short mitochondrial form of p19ARF induces autophagy and caspase-independent cell death. Mol Cell. 2006;22:463–75. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous