A Drosophila pasha mutant distinguishes the canonical microRNA and mirtron pathways - PubMed (original) (raw)
A Drosophila pasha mutant distinguishes the canonical microRNA and mirtron pathways
Raquel Martin et al. Mol Cell Biol. 2009 Feb.
Abstract
Canonical primary microRNA (miRNA) transcripts and mirtrons are proposed to transit distinct nuclear pathways en route to generating mature approximately 22 nucleotide regulatory RNAs. We generated a null allele of Drosophila pasha, which encodes a double-stranded RNA-binding protein partner of the RNase III enzyme Drosha. Analysis of this mutant yielded stringent evidence that Pasha is essential for the biogenesis of canonical miRNAs but is dispensable for the processing and function of mirtron-derived regulatory RNAs. The pasha mutant also provided a unique tool to study the developmental requirements for Drosophila miRNAs. While pasha adult somatic clones are similar in many respects to those of dicer-1 clones, pasha mutant larvae revealed an unexpected requirement for the miRNA pathway in imaginal disc growth. These data suggest limitations to somatic clonal analysis of miRNA pathway components.
Figures
FIG. 1.
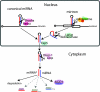
Canonical miRNA and mirtron pathways in Drosophila. Key protein families include RNase III endonucleases (Drosha and Dicer-1), double-stranded RNA-binding domain proteins (Pasha and Loqs) and Argonaute effectors (AGO1 and AGO2). Canonical miRNA precursors are cleaved by the Drosha/Pasha complex in the nucleus, cleaved again by the Dicer-1/Loqs complex in the cytoplasm, and predominantly loaded into AGO1. Mirtrons are short hairpin introns that use the splicing and debranching machinery to bypass the requirement for Drosha/Pasha but are subsequently processed by Dicer-1 to generate miRNA-class regulatory RNAs.
FIG. 2.
Scheme for the generation of the pasha KO deletion allele. (A) FLP-mediated recombination was used to delete genomic sequence between FRT-bearing piggyBac elements that flank the pasha locus. (B) The progenitor chromosomes bear piggyBac insertion in trans, which were brought in cis as a hybrid element after recombination. (C) PCR analysis verifies that the pasha KO allele juxtaposes the left (L) and right (R) arms of the progenitor piggyBacs, while presence of the novel hybrid (H) product reflects the deletion of the intervening pasha locus. (D) RT-PCR tests demonstrate that pasha transcripts are absent from pasha KO larvae, while expression of the neighboring locus CG1792 is maintained. Quantitative tests indicate 1.65-fold increase in CG1792 mRNA in this mutant (see Fig. S1 in the supplemental material).
FIG. 3.
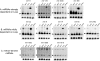
Small RNA expression in mutants for canonical miRNA and RNAi factors. Total RNAs were extracted from homozygous larvae of the following genotypes: Canton S (CS) = wild type, ago2 = ago2 414; dcr2 = dcr-2 L811fs; loqs = loqs KO; pasha = pasha KO. Blots were stripped and probed for 2S rRNA as a loading and transferring control (shown beneath each miRNA blot). Some blots were probed with multiple miRNAs and therefore have the same 2S control. (A) Canonical miRNAs that are strongly dependent on Loqs; i.e., for which the mature species is reduced (arrowhead) and there is accumulation of pre-miRNA hairpins (bracket). All of these show reduced miRNA and pre-miRNA levels in pasha KO. (B) miRNAs that are mildly dependent on Loqs; i.e., for which there is pre-miRNA accumulation but mature species are relatively unaffected. All of these still show reduced miRNA and pre-miRNA in pasha. (C) Mirtron-derived miRNAs. These are strongly affected in loqs but mostly unaffected in pasha. There is a slight reduction in pre-mir-1010 and miR-1010 in pasha, although this is potentially due to an effect on the expression of its host gene CG31163.
FIG. 4.
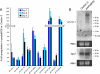
Pasha KO larvae strongly accumulate pri-miRNA species. (A) qRT-PCR) was used to assess pri-miRNA levels in Canton S and homozygous pasha KO larvae, as normalized to rp49. Three independent RNA samples were reverse transcribed for each genotype, and six qPCR measurements were made for each cDNA preparation. The y axis depicts the pri-miRNA ratio and standard error determined for each biological replicate. (B) Northern analysis of pri-mir-1 in Canton S and homozygous pasha KO larvae revealed the accumulation of an ∼8-kb transcript in the pasha mutant. Hybridization with an ago2 probe and ethidium staining of rRNA served as loading controls.
FIG. 5.
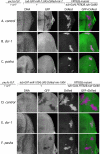
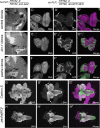
pasha distinguishes the function of canonical miRNAs and mirtrons. Shown are eye imaginal discs stained for DNA and a GFP sensor, with DsRed fluorescence marking active small RNA transgenes; the right panels depict the merged GFP/DsRed channels. (A to C) MARCM clones that are homozygous for a given chromosome 3R and ectopically express a hybrid DsRed:mir-7 transgene were tested for their ability to repress a miRNA sensor, a ubiquitously transcribed GFP target bearing two miR-7 binding sites. miRNA-mediated target repression was detected in control clones (A), but not in dcr-1 clones (B) or pasha KO clones (C). (D to F) MARCM clones that are homozygous for a given chromosome 3R and ectopically express a hybrid DsRed:mir-1004 (mirtron) transgene were tested for their ability to repress a mirtron sensor, a ubiquitously transcribed GFP transcript bearing two miR-1004 binding sites. Mirtron-mediated target repression was detected in control clones (D) but not dcr-1 clones (E); pasha KO homozygous cells generated active mirtrons (F).
FIG. 6.
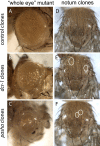
Phenotypes of dcr-1 and pasha KO adult mutant clones. Compared to wild-type, clonal loss of either Dcr-1 or Pasha results in small rough eyes (A to C) and loss of external mechanosensory bristle structures (D to F). Eye clones were made using the EGUF system (49), which generates eyes that are nearly composed entirely of mutant tissue.
FIG. 7.
Distinct growth defects observed in mutant clones compared to homozygous pasha KO animals. (A to C) Small retinal clones in third instar eye imaginal discs stained with β-galactosidase antibody. Homozygous mutant cells have no β-galactosidase (−/−), their twin-spots have a high level of β-galactosidase (+/+), and unrecombined heterozygous cells have an intermediate level of β-galactosidase (+/−). Compared to control clones, dcr-1 and pasha KO clones are much smaller than their respective twin-spots. (D to F) Large clones in third-instar eye imaginal discs generated with the Minute technique and stained with GFP antibody; the magnification is lower than in panels A to C. Minute homozygosity is cell lethal, and Minute heterozygous cells have a severe growth disadvantage, thus permitting the recovery of large, GFP−/− mutant clones. (D) Control clones occupy nearly the entire disc; the arrow and arrowhead point to small patches of heterozygous cells in the antenna and retina, respectively. Homozygous dcr-1 cells (E) and pasha KO (F) cells compete poorly even against Minute cells, so large GFP+ sectors remain. (G) Wild-type brain/imaginal disc complex stained for the neural marker Elav; the olfactory lobe (OL), ventral ganglion (VG), eye-antennal disc (E), and leg disc (L) are indicated. Elav is highly expressed by the brain and the developing retina of the eye disc (arrows, G′). (H) Brain/imaginal disc complex from a homozygous pasha KO larva is nearly devoid of imaginal discs and differentiate only small patches of Elav+ retina. The pasha KO brain exhibits rudimentary optic lobes, but the ventral ganglion is of fairly normal size.
Similar articles
- MicroRNA-dependent roles of Drosha and Pasha in the Drosophila larval ovary morphogenesis.
Yang H, Li M, Hu X, Xin T, Zhang S, Zhao G, Xuan T, Li M. Yang H, et al. Dev Biol. 2016 Aug 15;416(2):312-23. doi: 10.1016/j.ydbio.2016.06.026. Epub 2016 Jun 20. Dev Biol. 2016. PMID: 27339292 - Genome-wide identification of targets of the drosha-pasha/DGCR8 complex.
Kadener S, Rodriguez J, Abruzzi KC, Khodor YL, Sugino K, Marr MT 2nd, Nelson S, Rosbash M. Kadener S, et al. RNA. 2009 Apr;15(4):537-45. doi: 10.1261/rna.1319309. Epub 2009 Feb 17. RNA. 2009. PMID: 19223442 Free PMC article. - A Drosophila genetic screen yields allelic series of core microRNA biogenesis factors and reveals post-developmental roles for microRNAs.
Smibert P, Bejarano F, Wang D, Garaulet DL, Yang JS, Martin R, Bortolamiol-Becet D, Robine N, Hiesinger PR, Lai EC. Smibert P, et al. RNA. 2011 Nov;17(11):1997-2010. doi: 10.1261/rna.2983511. Epub 2011 Sep 23. RNA. 2011. PMID: 21947201 Free PMC article. - Mirtrons: microRNA biogenesis via splicing.
Westholm JO, Lai EC. Westholm JO, et al. Biochimie. 2011 Nov;93(11):1897-904. doi: 10.1016/j.biochi.2011.06.017. Epub 2011 Jun 21. Biochimie. 2011. PMID: 21712066 Free PMC article. Review. - The long and short of inverted repeat genes in animals: microRNAs, mirtrons and hairpin RNAs.
Okamura K, Chung WJ, Lai EC. Okamura K, et al. Cell Cycle. 2008 Sep 15;7(18):2840-5. doi: 10.4161/cc.7.18.6734. Epub 2008 Sep 5. Cell Cycle. 2008. PMID: 18769156 Free PMC article. Review.
Cited by
- The Drosophila Dicer-1 Partner Loquacious Enhances miRNA Processing from Hairpins with Unstable Structures at the Dicing Site.
Lim MY, Ng AW, Chou Y, Lim TP, Simcox A, Tucker-Kellogg G, Okamura K. Lim MY, et al. Cell Rep. 2016 May 24;15(8):1795-808. doi: 10.1016/j.celrep.2016.04.059. Epub 2016 May 12. Cell Rep. 2016. PMID: 27184838 Free PMC article. - MicroRNA biogenesis: regulating the regulators.
Finnegan EF, Pasquinelli AE. Finnegan EF, et al. Crit Rev Biochem Mol Biol. 2013 Jan-Feb;48(1):51-68. doi: 10.3109/10409238.2012.738643. Epub 2012 Nov 19. Crit Rev Biochem Mol Biol. 2013. PMID: 23163351 Free PMC article. Review. - Neural specificity of the RNA-binding protein Elav is achieved by post-transcriptional repression in non-neural tissues.
Sanfilippo P, Smibert P, Duan H, Lai EC. Sanfilippo P, et al. Development. 2016 Dec 1;143(23):4474-4485. doi: 10.1242/dev.141978. Epub 2016 Oct 17. Development. 2016. PMID: 27802174 Free PMC article. - Drosophila Argonaute 1 and its miRNA biogenesis partners are required for oocyte formation and germline cell division.
Azzam G, Smibert P, Lai EC, Liu JL. Azzam G, et al. Dev Biol. 2012 May 15;365(2):384-94. doi: 10.1016/j.ydbio.2012.03.005. Epub 2012 Mar 14. Dev Biol. 2012. PMID: 22445511 Free PMC article. - [Diversity and complexity of dicer dependent small RNA networks in animals].
Okamura K, Lai EC. Okamura K, et al. Seikagaku. 2009 Oct;81(10):904-9. Seikagaku. 2009. PMID: 19928532 Free PMC article. Review. No abstract available.
References
- Brennecke, J., D. R. Hipfner, A. Stark, R. B. Russell, and S. M. Cohen. 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 11325-36. - PubMed
- Carmell, M. A., and G. J. Hannon. 2004. RNase III enzymes and the initiation of gene silencing. Nat. Struct. Mol. Biol. 11214-218. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM083300/GM/NIGMS NIH HHS/United States
- R01 GM083300-02/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01-GM083300/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials