p53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest - PubMed (original) (raw)
p53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest
Christian J Braun et al. Cancer Res. 2008.
Abstract
microRNAs provide a novel layer of regulation for gene expression by interfering with the stability and/or translation of specific target mRNAs. Overall levels of microRNAs are frequently down-regulated in cancer cells, and reducing general microRNA processing increases cancerogenesis in transgenic models, suggesting that at least some microRNAs might act as effectors in tumor suppression. Accordingly, the tumor suppressor p53 up-regulates miR-34a, a microRNA that contributes to apoptosis and acute senescence. Here, we used array hybridization to find that p53 induces two additional, mutually related clusters of microRNAs, leading to the up-regulation of miR-192, miR-194, and miR-215. The same microRNAs were detected at high levels in normal colon tissue but were severely reduced in many colon cancer samples. On the other hand, miR-192 and its cousin miR-215 can each contribute to enhanced CDKN1A/p21 levels, colony suppression, cell cycle arrest, and cell detachment from a solid support. These effects were partially dependent on the presence of wild-type p53. Antagonizing endogenous miR-192 attenuated 5-fluorouracil-induced accumulation of p21. Hence, miR-192 and miR-215 can act as effectors as well as regulators of p53; they seem to suppress cancerogenesis through p21 accumulation and cell cycle arrest.
Conflict of interest statement
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Figures
Figure 1
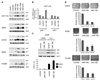
microRNA expression profiling. RNA samples enriched for small RNA molecules, extracted from SJSA cells treated with 8 µmol/L Nutlin-3 or DMSO alone, were conjugated with Cy3 or Cy5 dyes and hybridized to three microarrays. A dye swap was performed on another three microarrays. A, the raw signal level for each probe is presented in relative fluorescence units (r. fl.), calculated as an average from three microarrays (see Supplementary Table S2 for raw signal values). Rhombi represent all analyzed microRNAs, triangles represent spiked-in synthetic control RNA molecules, and crossed squares represent human microRNAs that showed a notable differential expression. B, fluorescent signals of spots corresponding to human microRNAs with a relative fluorescence over 1,000 were normalized according to let-7 gene family members. The relative induction is presented for each microRNA. Six microarrays, including dye swap, were analyzed. Columns, mean for each microRNA; bars, SE. The names of microRNAs represented by the columns correspond to the order shown in Supplementary Table S1.
Figure 2
Induction of microRNA expression through p53. A, qRT-PCR analysis of miR-34a, miR-192, miR-194, and miR-215 in SJSA, HCT116 p53 wt, and p53−/−, as well as U2OS cells treated with 8 µmol/L Nutlin-3 for 24 h. Fifty nanograms of extracted RNA enriched for small RNA molecules were subjected to qRT-PCR to quantify miR-34a, miR-192, miR-194, and miR-215, with miR-16 used for internal normalization. Columns, relative microRNA expression levels (“fold induction”) as calculated from the results of three independent experiments; bars, SE. B, SJSA cells (p53 wt) were treated with the doxorubicin, and the relative levels of microRNAs (fold induction) were then determined by qRT-PCR. A549 (p53 wt) or DLD-1 (p53 mutant) were treated with the indicated drugs [camptothecin (CPT), 200 nmol/L; doxorubicin (DOX), 100 ng/mL] and/or siRNA as detailed in Materials and Methods. The relative levels of miR-192 (fold induction) were then determined by qRT-PCR. C, presumptive p53-binding site on chromosome 1, 2.7 kb upstream of the miR-194-1/miR-215 cluster, with comparison with the known consensus sequence bound by p53 (43). D, ChIP of p53 with the genomic fragment comprising the sequence element shown in C. A549 cells were treated with camptothecin (300 nmol/L) for 24 h. Cells were harvested and then subjected to ChIP analysis with antibodies directed against p53 or the HA tag as a control. Precipitated DNA was analyzed by qPCR with primers amplifying the known 5′ p53-binding site of the CDKN1A/p21 promoter and the putative binding site near the miR-194-1/miR-215 gene cluster. A fragment of the MT-RNR2 gene was used as negative control. Columns, percentage of precipitated DNA compared with the amount of input. The results from two independently performed experiments are shown in each case, with brackets below the columns representing equivalent conditions.
Figure 3
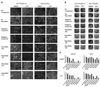
miR-192 levels in colon and colorectal cancer. A, RNA samples were extracted from different colorectal cancer cell lines (HCT116, Lovo, HT29, DLD-1, and HCC-2998) and also from the lung adenocarcinoma cell line A549. miR-192 levels were quantified by qRT-PCR as described in Materials and Methods. B, RNA samples were extracted from HCT116 p53 wt or p53−/− cells. miR-192 levels were quantified by qRT-PCR as described in Materials and Methods. C, microRNA expression levels from patient material were determined by array hybridization. The log2 of hybridization intensities were determined for each sample, grouped according to its origin: normal colon mucosa, colon cancer with MSI, and colon cancer with MSS, as shown in details in Supplementary Fig. S3 and Supplementary Tables S2 to S4. The data from this study are summarized in a box plot format, including the level of significance that distinguishes the MSS tumors from normal tissue for each microRNA.
Figure 4
p21 induction and colony suppression by p53-responsive microRNAs. A, cells of the indicated lines were transfected with the indicated microRNAs (10 nmol/L) and incubated for 48 h; then, cell lysates were subjected to immunoblot analysis to detect p53, p21, or β-actin (input control). B, HCT116 cells, p53 wt or p53−/−, were transfected with either control microRNAs or miR-192 (10 nmol/L) and incubated for 48 h, and p21 mRNA levels were quantified by qRT-PCR. Columns, mean values obtained from three independent experiments, normalized to the mock-transfected p53 wt cells; bars, SE. C, A549 cells were transfected with either scrambled LNA or anti-miR-192 LNA (100 nmol/L) and then (24 h after transfection) treated with 500 µmol/L 5-FU or the DMSO solvent alone. After 24 h of 5-FU treatment, the cells were harvested and cell lysates were subjected to immunoblot analysis. In parallel, flow cytometry was performed after transfection and 48 h of incubation with 500 µmol/L 5-FU or DMSO alone. The cells were then trypsinized, fixed with ethanol, and stained with propidium iodide. The percentages of cells with a DNA content below 2N (sub-G1, gated to the total number of cells) are indicated in each case. The columns are based on the raw data shown in Supplementary Fig. S5 and indicate the mean values obtained from three independent experiments; bars, SE. D, U2OS, H1299, or SJSA cells were transfected with control miR-vector or miR-vec-34a or miR-vec-192. The cultures were maintained for 2 wk with blasticidin (5 µg/mL) to select stable transfectants, and the cells were then fixed, stained with crystal violet, and photographed. The numbers of colonies were determined by a person unaware of the identity of the samples, and the results from three experiments are shown in the column diagrams. Columns, mean; bars, SE.
Figure 5
Cell cycle arrest and senescence induced by p53-responsive microRNAs. A, extent of apoptosis after transfection of HCT116 p53 wt or p53−/− cells with the indicated pre-miR molecules and 48 h of incubation. The cells were trypsinized, fixed with ethanol, and stained with propidium iodide. The percentages of cells with a DNA content below 2N (sub-G1, gated to the total number of cells) are indicated in each case. An example of the raw data obtained by flow cytometry is shown in Supplementary Fig. S6. B, cells were prepared as in A. The diagrams indicate the percentage of cells in G1, S, and G2 phases of the cell cycle after transfection with the indicated pre-miR molecules. Flow cytometry diagrams are shown in Supplementary Fig. S6. C, HCT116 p53 wt or p53−/− cells transfected with 10 nmol/L of the indicated microRNAs were incubated for 2 d, and cells were then treated with nocodazole (100 ng/mL) for another 18 h. Cell cycle profiles were monitored as described in Materials and Methods. The diagram shows the percentages of cells with a DNA content below 2N (sub-G1, gated to the total number of cells). D, microRNAs were transfected as pre-miR molecules in HCT116 p53 wt cells. One well was left without transfection (no transfection) as a negative control, and one well was treated with low-dose camptothecin treatment (20 nmol/L) as a positive control. SA-β-Gal was stained after 4 d of incubation. Blue staining intensity of three pictures for every condition was estimated by Adobe Photoshop software. Staining intensity was normalized to miR-34a staining intensity.
Figure 6
Cell detachment induced by miR-192 and miR-215. HCT116 cells with or without a targeted disruption of TP53 were seeded at 60,000 per well in six-well plates and transfected with pre-miR molecules to express miR-34a, miR-192, miR-194, and miR-215. In addition, two molecules that are not occurring in nature were transfected (Negative Control #1 and Negative Control #2), and one well for each condition was treated with transfection reagent but without transfected nucleic acid (No MicroRNA) and one was cultured without nucleic acid and without transfection reagent (No Transfection). One day after transfection, camptothecin was added to a final concentration of 20 nmol/L, and negative controls were performed with DMSO alone. A, images were taken to show cell morphology after 48 h of incubation. B, cells were stained with crystal violet solution after another 48 h of camptothecin or DMSO incubation. Images were taken to show the entire wells and the remaining cells within them. C, blue pixel rates of the images shown in Fig. 4_B_ were determined using the Photoshop software and are indicated by columns for each condition.
Similar articles
- Mechanism of chemoresistance mediated by miR-140 in human osteosarcoma and colon cancer cells.
Song B, Wang Y, Xi Y, Kudo K, Bruheim S, Botchkina GI, Gavin E, Wan Y, Formentini A, Kornmann M, Fodstad O, Ju J. Song B, et al. Oncogene. 2009 Nov 19;28(46):4065-74. doi: 10.1038/onc.2009.274. Epub 2009 Sep 7. Oncogene. 2009. PMID: 19734943 Free PMC article. - MiR-1 Suppresses Proliferation of Osteosarcoma Cells by Up-regulating p21 via PAX3.
Fujii R, Osaka E, Sato K, Tokuhashi Y. Fujii R, et al. Cancer Genomics Proteomics. 2019 Jan-Feb;16(1):71-79. doi: 10.21873/cgp.20113. Cancer Genomics Proteomics. 2019. PMID: 30587501 Free PMC article. - Tumor suppressor miR-22 determines p53-dependent cellular fate through post-transcriptional regulation of p21.
Tsuchiya N, Izumiya M, Ogata-Kawata H, Okamoto K, Fujiwara Y, Nakai M, Okabe A, Schetter AJ, Bowman ED, Midorikawa Y, Sugiyama Y, Aburatani H, Harris CC, Nakagama H. Tsuchiya N, et al. Cancer Res. 2011 Jul 1;71(13):4628-39. doi: 10.1158/0008-5472.CAN-10-2475. Epub 2011 May 12. Cancer Res. 2011. PMID: 21565979 Free PMC article. - Roles of TP53 in determining therapeutic sensitivity, growth, cellular senescence, invasion and metastasis.
McCubrey JA, Lertpiriyapong K, Fitzgerald TL, Martelli AM, Cocco L, Rakus D, Gizak A, Libra M, Cervello M, Montalto G, Yang LV, Abrams SL, Steelman LS. McCubrey JA, et al. Adv Biol Regul. 2017 Jan;63:32-48. doi: 10.1016/j.jbior.2016.10.001. Epub 2016 Oct 6. Adv Biol Regul. 2017. PMID: 27776972 Review. - P53/microRNA-34-induced metabolic regulation: new opportunities in anticancer therapy.
Zhang DG, Zheng JN, Pei DS. Zhang DG, et al. Mol Cancer. 2014 May 21;13:115. doi: 10.1186/1476-4598-13-115. Mol Cancer. 2014. PMID: 24884974 Free PMC article. Review.
Cited by
- Role of Angiogenesis and Its Biomarkers in Development of Targeted Tumor Therapies.
Pathak A, Pal AK, Roy S, Nandave M, Jain K. Pathak A, et al. Stem Cells Int. 2024 Jan 4;2024:9077926. doi: 10.1155/2024/9077926. eCollection 2024. Stem Cells Int. 2024. PMID: 38213742 Free PMC article. Review. - Noncoding RNAs in DNA repair and genome integrity.
Wan G, Liu Y, Han C, Zhang X, Lu X. Wan G, et al. Antioxid Redox Signal. 2014 Feb 1;20(4):655-77. doi: 10.1089/ars.2013.5514. Epub 2013 Sep 18. Antioxid Redox Signal. 2014. PMID: 23879367 Free PMC article. Review. - Transcription poisoning by Topoisomerase I is controlled by gene length, splice sites, and miR-142-3p.
Solier S, Ryan MC, Martin SE, Varma S, Kohn KW, Liu H, Zeeberg BR, Pommier Y. Solier S, et al. Cancer Res. 2013 Aug 1;73(15):4830-9. doi: 10.1158/0008-5472.CAN-12-3504. Epub 2013 Jun 20. Cancer Res. 2013. PMID: 23786772 Free PMC article. - Non-coding RNAs in DNA damage and repair.
Sharma V, Misteli T. Sharma V, et al. FEBS Lett. 2013 Jun 27;587(13):1832-9. doi: 10.1016/j.febslet.2013.05.006. Epub 2013 May 16. FEBS Lett. 2013. PMID: 23684639 Free PMC article. Review. - Astrocytes Can Adopt Endothelial Cell Fates in a p53-Dependent Manner.
Brumm AJ, Nunez S, Doroudchi MM, Kawaguchi R, Duan J, Pellegrini M, Lam L, Carmichael ST, Deb A, Hinman JD. Brumm AJ, et al. Mol Neurobiol. 2017 Aug;54(6):4584-4596. doi: 10.1007/s12035-016-9974-3. Epub 2016 Jul 8. Mol Neurobiol. 2017. PMID: 27389775 Free PMC article.
References
- Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. - PubMed
- Moll UM, Wolff S, Speidel D, Deppert W. Transcription-independent pro-apoptotic functions of p53. Curr Opin Cell Biol. 2005;17:631–636. - PubMed
- Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. - PubMed
- Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. - PubMed
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous