A stepwise pathway for biogenesis of 24-nt secondary siRNAs and spreading of DNA methylation - PubMed (original) (raw)
A stepwise pathway for biogenesis of 24-nt secondary siRNAs and spreading of DNA methylation
Lucia Daxinger et al. EMBO J. 2009.
Abstract
We used a transgene system to study spreading of RNA-directed DNA methylation (RdDM) during transcriptional gene silencing in Arabidopsis thaliana. Forward and reverse genetics approaches using this system delineated a stepwise pathway for the biogenesis of secondary siRNAs and unidirectional spreading of methylation from an upstream enhancer element into downstream sequences. Trans-acting, hairpin-derived primary siRNAs induce primary RdDM, independently of an enhancer-associated 'nascent' RNA, at the target enhancer region. Primary RdDM is a key step in the pathway because it attracts the secondary siRNA-generating machinery, including RNA polymerase IV, RNA-dependent RNA polymerase2 and Dicer-like3 (DCL3). These factors act in a turnover pathway involving a nascent RNA, which normally accumulates stably in non-silenced plants, to produce cis-acting secondary siRNAs that induce methylation in the downstream region. The identification of DCL3 in a forward genetic screen for silencing-defective mutants demonstrated a strict requirement for 24-nt siRNAs to direct methylation. A similar stepwise process for spreading of DNA methylation may occur in mammalian genomes, which are extensively transcribed in upstream regulatory regions.
Figures
Figure 1
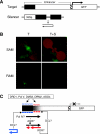
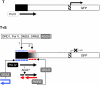
Transgene silencing system and hypothetical model for spreading of DNA methylation through secondary siRNAs. (A) The target locus (T) contains a meristem-specific enhancer (region targeted for methylation shown in black) placed upstream of a minimal promoter (hatched) and _GFP_-coding region. The unlinked silencer locus (S) contains a transcribed inverted DNA repeat of distal enhancer sequences encoding a hairpin RNA. The hairpin RNA trigger is diced into short interfering (si) RNAs, which induce de novo methylation of the target enhancer in trans. There is a short tandem repeat in the target enhancer (Kanno et al, 2008), which has an unknown function in spreading of methylation and silencing. 35Sp: 35S promoter of cauliflower mosaic virus. (B) Transgenic seedlings containing only the target locus (T) display GFP fluorescence in root and shoot apical meristem regions (RAM and SAM, respectively) (left); in seedlings containing the target and silencer loci (T+S), GFP fluorescence is abolished (right). (C) In the hypothetical model tested in this study, primary siRNAs (blue dashes) originating from DCL processing of the hairpin RNA trigger induce primary RdDM (blue ‘m') of the targeted enhancer region (black box) and may guide AGO slicing of an overlapping nascent RNA (black arrow), which is proposed to be transcribed by Pol IV from the methylated DNA template. The Pol IV-generated nascent RNA, or cleavage fragments thereof, provide substrates for RDR; the resulting double-stranded RNA is diced to generate 24-nt secondary siRNAs (red dashes) that induce secondary RdDM (red ‘m') in the downstream region (black shade). GFP expression is silenced. Primary and secondary RdDM require DRD1, Pol V and DMS3 (Kanno et al, 2008) and are presumably catalysed by the de novo methyltransferase DRM2 (Cao et al, 2003) with the participation of AGO4 (Chan et al, 2004).
Figure 2
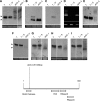
RNA profiles in rdr2, rdr6, nrpd1 and ago4 mutants. Northern blots of small RNAs: (A) primary siRNAs derived from hairpin RNA trigger; (B) secondary siRNAs; (C) siRNA02; (D) tasiRNA255. The arrowheads at the right indicate 21- and 24-nt-size classes. Ethidium bromide staining of the major RNA on the gels is shown as a loading control. The same blot was reprobed in B–D. (E) RT–PCR detection of nascent RNA; (F) RT–PCR of an actin control; (G) GFP expression. The DNA lane is amplified genomic DNA. Abbreviations: T/T;−/−, non-silenced plants containing a homozygous target locus; T/T;S/(S), silenced plants containing a homozygous target locus and either a hemizygous or homozygous silencer locus; WT, wild-type transgenic plant; Col-0, wild-type non-transgenic plants. For each of the four mutants tested (small letters), results from wild-type siblings (capital letters) are shown for comparison. The quantitative differences in primary siRNAs (A) are most likely due to the fact that RNA was prepared from pools of F2 or F3 plants that could be either homozygous or hemizygous for the silencer locus.
Figure 3
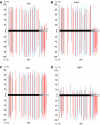
Analysis of DNA methylation at the targeted enhancer and downstream region by bisulphite sequencing in rdr2, rdr6, nrpd1 and ago4 mutants. The graphs show the percentage of methylation at individual cytosines in mutant plants (top) and their wild-type siblings (bottom). (A) rdr2/RDR2; (B) nrpd1/NRPD1; (C) rdr6/RDR6; (D) ago4/AGO4. The black bar represents the enhancer region targeted by primary siRNAs (primary RdDM); the shaded grey bar indicates the downstream region targeted by secondary siRNAs (secondary RdDM). Black lines: CG methylation; blue lines: CNG methylation; red lines: CNN methylation. Residual secondary RdDM in rdr2 may be due to low levels of 24-nt siRNAs produced by RDR6/DCL3 activities (Gasciolli et al, 2005; Eamens et al, 2008). The results are from at least 10 cloned sequences. Original data are shown in Supplementary Figure 4.
Figure 4
RNA profile in the dcl3-5 mutant. Northern blots of small RNAs (A–C; E–J) and RT–PCR detection of the nascent RNA (D). (A) siRNA1003; (B) primary siRNAs; (C) secondary siRNAs; (D) nascent RNA (top) and actin control (bottom); (E) 45S rDNA siRNAs; (F) siRNA02; (G) solo LTR siRNAs; (H) Tag2 siRNAs; (I) Sat5 siRNAs. The arrowheads at the left indicate 21- and 24-nt size classes. In panels G–I, the lanes were pieced together from the same gel, as indicated by the dividing white lines. At the bottom of each blot, ethidium bromide staining of the major RNA on the gels is shown as a loading control. Bottom: domain structure of the DCL3 protein (1531 amino acids; Schauer et al, 2002) and position of the dcl3-5 mutation. It is unclear why a dcl3 mutation was recovered in this mutant screen and not a previous one based on silencing a seed-specific promoter (Kanno et al, 2004, 2005), but it may reflect the enhanced sensitivity of the present screen, which uses detection of enhanced GFP in colourless root tips.
Figure 5
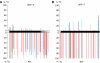
Analysis of DNA methylation at the targeted enhancer and an endogenous Tag2 element by bisulphite sequencing in the dcl3-5 mutant. The graphs show the percentage of methylation at individual cytosines in the dcl3-5 mutant (top) and wild-type plants (bottom) in the following sequences. (A) Targeted enhancer region (black bar) and downstream region (shaded grey bar); (B) Tag2 element. Black lines: CG methylation; blue lines: CNG methylation; red lines: CNN methylation. The results are from eight cloned sequences. Original data are shown in Supplementary Figure 4. The nearly complete loss of asymmetric CNN methylation in both sequences indicates a defect in de novo methylation; the residual CG and CNG methylation is probably due to siRNA-independent maintenance of symmetrical methylation (Aufsatz et al, 2002).
Figure 6
Stepwise pathway for secondary siRNA biogenesis and spreading of methylation. Proteins whose functions have been demonstrated in this study are shown in white letters on dark boxes. Top: in non-silenced plants containing the target locus (T) only, the targeted enhancer region (black box) is unmethylated and the nascent RNA, which normally does not interfere with GFP expression, is presumably transcribed by Pol II because it is still detectable in an nrpd1 mutant. Bottom: in the presence of the silencer locus (S), GFP expression is silenced by primary RdDM (blue ‘m'), which is induced by hairpin-derived, DCL3-dependent 24-nt primary siRNAs (blue dashes). Primary RdDM attracts the secondary siRNA-generating machinery, which includes Pol IV, RDR2 and DCL3. In this model, Pol IV transcribes the methylated template to produce the nascent RNA, which is copied and diced by RDR2 and DCL3, respectively, to produce 24-nt secondary siRNAs (red dashes) that guide secondary RdDM (red ‘m') in the downstream region (black shade). Pol IV may also transcribe the double-stranded RNA to sustain secondary siRNA production once the cycle is initiated (Supplementary Figure 2). In both cases, Pol IV transcribes a precursor of secondary siRNAs. Whether AGO cleavage of the nascent RNA is required to provide substrates for RDR2 is still not known. siRNA-guided de novo methylation requires DRD1, DMS3 and Pol V (Kanno et al, 2008) and is presumably catalysed by DRM2 (Cao et al, 2003).
Similar articles
- Distinct and concurrent pathways of Pol II- and Pol IV-dependent siRNA biogenesis at a repetitive trans-silencer locus in Arabidopsis thaliana.
Sasaki T, Lee TF, Liao WW, Naumann U, Liao JL, Eun C, Huang YY, Fu JL, Chen PY, Meyers BC, Matzke AJ, Matzke M. Sasaki T, et al. Plant J. 2014 Jul;79(1):127-38. doi: 10.1111/tpj.12545. Epub 2014 Jun 13. Plant J. 2014. PMID: 24798377 - Interplay among RNA polymerases II, IV and V in RNA-directed DNA methylation at a low copy transgene locus in Arabidopsis thaliana.
You W, Lorkovic ZJ, Matzke AJ, Matzke M. You W, et al. Plant Mol Biol. 2013 May;82(1-2):85-96. doi: 10.1007/s11103-013-0041-4. Epub 2013 Mar 20. Plant Mol Biol. 2013. PMID: 23512103 Free PMC article. - Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis.
Blevins T, Podicheti R, Mishra V, Marasco M, Wang J, Rusch D, Tang H, Pikaard CS. Blevins T, et al. Elife. 2015 Oct 2;4:e09591. doi: 10.7554/eLife.09591. Elife. 2015. PMID: 26430765 Free PMC article. - RNA-directed DNA methylation in plants: Where to start?
Zhang H, He X, Zhu JK. Zhang H, et al. RNA Biol. 2013 Oct;10(10):1593-6. doi: 10.4161/rna.26312. RNA Biol. 2013. PMID: 25003825 Free PMC article. Review. - Cell biology of the Arabidopsis nuclear siRNA pathway for RNA-directed chromatin modification.
Pikaard CS. Pikaard CS. Cold Spring Harb Symp Quant Biol. 2006;71:473-80. doi: 10.1101/sqb.2006.71.046. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381329 Review.
Cited by
- Primer-dependent and primer-independent initiation of double stranded RNA synthesis by purified Arabidopsis RNA-dependent RNA polymerases RDR2 and RDR6.
Devert A, Fabre N, Floris M, Canard B, Robaglia C, Crété P. Devert A, et al. PLoS One. 2015 Mar 20;10(3):e0120100. doi: 10.1371/journal.pone.0120100. eCollection 2015. PLoS One. 2015. PMID: 25793874 Free PMC article. - Generation of marker-free transgenic plants concurrently resistant to a DNA geminivirus and a RNA tospovirus.
Yang CF, Chen KC, Cheng YH, Raja JA, Huang YL, Chien WC, Yeh SD. Yang CF, et al. Sci Rep. 2014 Jul 17;4:5717. doi: 10.1038/srep05717. Sci Rep. 2014. PMID: 25030413 Free PMC article. - Atypical DNA methylation of genes encoding cysteine-rich peptides in Arabidopsis thaliana.
You W, Tyczewska A, Spencer M, Daxinger L, Schmid MW, Grossniklaus U, Simon SA, Meyers BC, Matzke AJ, Matzke M. You W, et al. BMC Plant Biol. 2012 Apr 19;12:51. doi: 10.1186/1471-2229-12-51. BMC Plant Biol. 2012. PMID: 22512782 Free PMC article. - RNA-directed DNA methylation and demethylation in plants.
Chinnusamy V, Zhu JK. Chinnusamy V, et al. Sci China C Life Sci. 2009 Apr;52(4):331-43. doi: 10.1007/s11427-009-0052-1. Epub 2009 Apr 21. Sci China C Life Sci. 2009. PMID: 19381459 Free PMC article. Review. - Functional divergence in the growing family of RNA polymerases.
Landick R. Landick R. Structure. 2009 Mar 11;17(3):323-5. doi: 10.1016/j.str.2009.02.006. Structure. 2009. PMID: 19278646 Free PMC article.
References
- Bao N, Lye KW, Barton MK (2004) MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev Cell 7: 653–662 - PubMed
- Bei Y, Pressman S, Carthew R (2007) SnapShot: small RNA-mediated epigenetic modifications. Cell 130: 756. - PubMed
- Blewitt M, Gendrel AV, Pang Z, Sparrow DB, Whitelaw N, Craig JM, Apedaile A, Hilton DJ, Dunwoddie SL, Brockdorff N, Kay GF, Whitelaw E (2008) SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat Genet 40: 663–669 - PubMed
- Cao X, Aufsatz W, Zilberman D, Mette MF, Huang MS, Matzke M, Jacobsen SE (2003) Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol 13: 2212–2217 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials