PolV(PolIVb) function in RNA-directed DNA methylation requires the conserved active site and an additional plant-specific subunit - PubMed (original) (raw)
PolV(PolIVb) function in RNA-directed DNA methylation requires the conserved active site and an additional plant-specific subunit
Sylvie Lahmy et al. Proc Natl Acad Sci U S A. 2009.
Abstract
Two forms of a plant-specific RNA polymerase (Pol), PolIV(PolIVa) and PolV(PolIVb), currently defined by their respective largest subunits [NRPD1(NRPD1a) and NRPE1(NRPD1b)], have been implicated in the production and activity of 24-nt small RNAs (sRNAs) in RNA-directed DNA methylation (RdDM). Prevailing models support the view that PolIV(PolIVa) plays an upstream role in RdDM by producing the 24-nt sRNAs, whereas PolV(PolIVb) would act downstream at a structural rather than an enzymatic level to reinforce sRNA production by PolIV(PolIVa) and mediate DNA methylation. However, the composition and mechanism of action of PolIV(PolIVa)/PolV(PolIVb) remain unclear. In this work, we have identified a plant-specific PolV(PolIVb) subunit, NRPE5a, homologous to NRPB5a, a common subunit shared by PolI-III and shown here to be present in PolIV(PolIVa). Our results confirm the combinatorial diversity of PolIV(PolIVa)/PolV(PolIVb) subunit composition and indicate that these plant-specific Pols are eukaryotic-type polymerases. Moreover, we show that nrpe5a-1 mutation differentially impacts sRNAs accumulation at various PolIV(PolIVa)/PolV(PolIVb)-dependent loci, indicating a target-specific requirement for NRPE5a in the process of PolV(PolIVb)-dependent gene silencing. We then describe that the triad aspartate motif present in the catalytic center of PolV(PolIVb) is required for recapitulation of all activities associated with this Pol complex in RdDM, suggesting that RNA polymerization is important for PolV(PolIVb) to perform its regulatory functions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
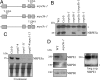
Fig. 1.
Higher plants contain 2 classes of RPB5-type Pol subunits. (A) Genes identified for each common Pol subunit in the Arabidopsis genome. Identical amino acid percentages between the Arabidopsis sequences are given for each universal subunit. (B) Diagrams show ScRPB5 and its homologues in Arabidopsis. The overall amino acid identity between NRPB5a and its homologues is indicated for the Jaw and the Assembly domains. AtNRPB5a and -b correspond to At3g22320 and At5g57980, respectively, whereas AtNRPE5a, -b, and -c correspond to At3g57080, At2g41340, and At3g55490. (C) Evolutionary relationships between NRPB5-type proteins and related factors. The unrooted phylogenetic tree was inferred from the full-length protein alignment from
Fig. S1
. Sc, Saccharomyces cerevisiae; At, Arabidopsis thaliana; Os, Oryza sativa; Hs, Homo sapiens; Dm, Drosophila melanogaster; Pf, Pyrococcus furiosus; Sa, Sulfolobus acidocaldarius. (D) RT-PCR analysis of NRPB5a and NRPE5s expression in various Arabidopsis tissues, including roots (Ro), leaves (Le), flower buds (FB), flowers (Fl), siliques (Si), and seeds (Se). EF1-4α was used as a control. Ge represents the genomic DNA control.
Fig. 2.
Mutual stability of NRPE5a and NRPE1(NRPD1b) in various null mutants. (A) Diagram of the _AtNRPE5_-type genes in Arabidopsis. Translated exons are indicated with open gray boxes. Vertical open arrowheads indicate the location of the T-DNA insertions. (B) Western blot analysis of total extracts from wild-type (Ws/Col-0) and independent nrpe5 single/double mutants using the anti-NRPE5a antibodies. A nonspecific cross-reacting band (indicated by an asterisk) was used as a loading control. (C) Stability of the NRPE5a protein in single nrpd1-4 and nrpde1-11 and corresponding double mutants. Coomassie blue staining was shown as a loading control. (D) Stability of the NRPE1(NRPD1b) subunits in nrpe5a-1 mutant. NRPB1, the largest subunit of PolII, and a nonspecific anti-NRPE1(NRPD1b) cross-reacting band (indicated by an asterisk) were used as loading controls. Long exp, longer exposure.
Fig. 3.
NRPE5a is a plant-specific subunit of PolV(PolIVb). Physical interaction between NRPE5a and PolV(PolIVb) (A) or PolIV(PolIVa) (B) detected by immunoprecipitation. Protein extracts prepared from either wild-type or complemented NRPE1(NRPD1b)/NRPD1(NRPD1a)-Flag transgenic lines were immunoprecipitated using anti-Flag M2 antibody, and the input/pellet fractions were analyzed by Western blot. Membranes were analyzed to detect the Flag epitope, NRPE1(NRPD1b), NRPE5a, and NRPB5a Pol subunits (A) or Flag epitope, NRPB1, NRPE5a, and NRPB5a Pol subunits (B).
Fig. 4.
Partial loss of methylation and silencing of RdDM targets in nrpe5a-1 mutant. (A) Southern blot analysis of 5S rDNA loci on genomic DNA digested with methylation-sensitive enzyme HpaII in wild-type (WS) and _nrpe5a_-1. (B) Partial loss of CNN methylation at the solo LTR locus in wild-type vs. _nrpe5a_-1 plants (Top). AluI-, DdeI-, and nondigested DNA (Input) were used as a template for PCRs using solo LTR primers. Derepression of the solo LTR-driven IG/LINE expression in the _nrpe5a_-1 mutant was assessed by semiquantitative RT-PCR using primers located in the IG/LINE. The constitutively expressed GAPA gene is used as control (Bottom). (C) Differential effect of NRPE5a loss on sRNA accumulation. Total RNA (30 μg) from inflorescences was successively probed with various sRNAs, U6, and miR-159 as loading controls.
Fig. 5.
PolV(PolIVb) active site requirement for RdDM. (A) Sequence alignments in the conserved box D region of various Pol large subunits. The substitution in the catalytic triad of nrpe1-3/drd3-3 mutant protein is indicated. (B) Western blot analysis of total extracts from wild-type, nrpe1-11, and nrpe1-3/drd3-3 plants with the anti-NRPE1(NRPD1b) antibody (Top). The corresponding Coomassie staining is shown as a loading control. Immunoprecipitation of NRPE5a using anti-NRPE1(NRPD1b) antibody in wild-type, nrpe1-11, and nrpe1-3/drd3-3 extracts followed by the Western blot analysis of eluates with anti-NRPE1(NRPD1b) and anti-NRPE5a antibodies (Bottom). (C) Loss of CNN methylation at the solo-LTR locus in wild-type, nrpe1-11, and nrpe1-3/drd3-3 plants. Real-time PCR analysis of IG/LINE expression in wild-type, nrpe1-11, and nrpe1-3/drd3-3 plants. (D) Analysis of sRNA accumulation by Northern blot done with 30 μg of total RNA from inflorescences from wild-type, nrpe1-11, and nrpe1-3/drd3-3 plants.
Similar articles
- PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis.
Mosher RA, Schwach F, Studholme D, Baulcombe DC. Mosher RA, et al. Proc Natl Acad Sci U S A. 2008 Feb 26;105(8):3145-50. doi: 10.1073/pnas.0709632105. Epub 2008 Feb 19. Proc Natl Acad Sci U S A. 2008. PMID: 18287047 Free PMC article. - High-temperature effect on genes engaged in DNA methylation and affected by DNA methylation in Arabidopsis.
Naydenov M, Baev V, Apostolova E, Gospodinova N, Sablok G, Gozmanova M, Yahubyan G. Naydenov M, et al. Plant Physiol Biochem. 2015 Feb;87:102-8. doi: 10.1016/j.plaphy.2014.12.022. Epub 2015 Jan 1. Plant Physiol Biochem. 2015. PMID: 25576840 - RETINOBLASTOMA-RELATED interactions with key factors of the RNA-directed DNA methylation (RdDM) pathway and its influence on root development.
León-Ruiz J, Espinal-Centeno A, Blilou I, Scheres B, Arteaga-Vázquez M, Cruz-Ramírez A. León-Ruiz J, et al. Planta. 2023 Apr 30;257(6):105. doi: 10.1007/s00425-023-04135-x. Planta. 2023. PMID: 37120771 - Plant-specific multisubunit RNA polymerase in gene silencing.
Lahmy S, Bies-Etheve N, Lagrange T. Lahmy S, et al. Epigenetics. 2010 Jan 1;5(1):4-8. doi: 10.4161/epi.5.1.10435. Epub 2010 Jan 26. Epigenetics. 2010. PMID: 20173420 Review. - Taking the Wheel - de novo DNA Methylation as a Driving Force of Plant Embryonic Development.
Markulin L, Škiljaica A, Tokić M, Jagić M, Vuk T, Bauer N, Leljak Levanić D. Markulin L, et al. Front Plant Sci. 2021 Oct 29;12:764999. doi: 10.3389/fpls.2021.764999. eCollection 2021. Front Plant Sci. 2021. PMID: 34777448 Free PMC article. Review.
Cited by
- DNA-dependent RNA polymerases in plants.
Yang DL, Huang K, Deng D, Zeng Y, Wang Z, Zhang Y. Yang DL, et al. Plant Cell. 2023 Sep 27;35(10):3641-3661. doi: 10.1093/plcell/koad195. Plant Cell. 2023. PMID: 37453082 Free PMC article. - Pol V produced RNA facilitates transposable element excision site repair in Arabidopsis.
Renken K, Mendoza SM, Diaz S, Slotkin RK, Hancock CN. Renken K, et al. MicroPubl Biol. 2023 May 16;2023:10.17912/micropub.biology.000793. doi: 10.17912/micropub.biology.000793. eCollection 2023. MicroPubl Biol. 2023. PMID: 37273575 Free PMC article. - Broad noncoding transcription suggests genome surveillance by RNA polymerase V.
Tsuzuki M, Sethuraman S, Coke AN, Rothi MH, Boyle AP, Wierzbicki AT. Tsuzuki M, et al. Proc Natl Acad Sci U S A. 2020 Dec 1;117(48):30799-30804. doi: 10.1073/pnas.2014419117. Epub 2020 Nov 16. Proc Natl Acad Sci U S A. 2020. PMID: 33199612 Free PMC article. - Long-Read cDNA Sequencing Enables a "Gene-Like" Transcript Annotation of Transposable Elements.
Panda K, Slotkin RK. Panda K, et al. Plant Cell. 2020 Sep;32(9):2687-2698. doi: 10.1105/tpc.20.00115. Epub 2020 Jul 9. Plant Cell. 2020. PMID: 32647069 Free PMC article. - Involvement of MEM1 in DNA demethylation in Arabidopsis.
Lu Y, Dai J, Yang L, La Y, Zhou S, Qiang S, Wang Q, Tan F, Wu Y, Kong W, La H. Lu Y, et al. Plant Mol Biol. 2020 Feb;102(3):307-322. doi: 10.1007/s11103-019-00949-0. Epub 2020 Jan 4. Plant Mol Biol. 2020. PMID: 31902068
References
- Ebright RH. RNA polymerase: Structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J Mol Biol. 2000;304:687–698. - PubMed
- Cramer P. Multisubunit RNA polymerases. Curr Opin Struct Biol. 2002;12:89–97. - PubMed
- Brodersen P, Voinnet O. The diversity of RNA silencing pathways in plants. Trends Genet. 2006;22:268–280. - PubMed
- Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6:24–35. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials