Targeting Mdm2 and Mdmx in cancer therapy: better living through medicinal chemistry? - PubMed (original) (raw)
Review
Targeting Mdm2 and Mdmx in cancer therapy: better living through medicinal chemistry?
Mark Wade et al. Mol Cancer Res. 2009 Jan.
Abstract
Genomic and proteomic profiling of human tumor samples and tumor-derived cell lines are essential for the realization of personalized therapy in oncology. Identification of the changes required for tumor initiation or maintenance will likely provide new targets for small-molecule and biological therapeutics. For example, inactivation of the p53 tumor suppressor pathway occurs in most human cancers. Although this can be due to frank p53 gene mutation, almost half of all cancers retain the wild-type p53 allele, indicating that the pathway is disabled by other means. Alternate mechanisms include deletion or epigenetic inactivation of the p53-positive regulator arf, methylation of the p53 promoter, or elevated expression of the p53 regulators Mdm2 and Mdmx. This review discusses current models of p53 regulation by Mdm2 and Mdmx and presents the rationale for design of future Mdmx-specific therapeutics based on our knowledge of its structure and biological functions.
Figures
Figure 1. Mdm2/Mdmx heterodimers are more effective p53 ubiquitin ligases than mdm2 homodimers
Mdm2 can homodimerize or heterodimerize with mdmx via RING/RING interaction (mdm2 and mdmx RING depicted, the remainder of the protein is omitted for clarity). It is proposed that the mdm2/mdmx heterodimer (upper scheme) provides the optimal structure for E2-dependent p53 ubiquitination, whereas mdm2 homodimerization (lower scheme) creates a suboptimal structure for p53 ubiquitination. In this schematic, the structural differences in homo- versus heterodimers lead to different positioning of the E2 enzyme relative to p53. However, alternative explanations upstream of E2 recruitment, such as optimal p53 binding in the heterodimer, or downstream of E2 recruitment, such as more effective targeting of p53 to the proteasome by the heterodimer, cannot be excluded.
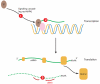
Figure 2. Blockade of mdmx at the level of transcription or translation
Inhibition of mitogen-activated signaling pathways (1) may prevent transcription factor recruitment to the mdmx promoter, thereby lowering mdmx mRNA. Disruption of transcription factor complexes (2) on the DNA could also reduce mdmx transcription. Degradation or reduced translation of mdmx mRNA following siRNA strategies (3) will also lead to decreased mdmx protein.
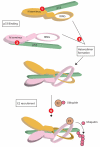
Figure 3. Mdmx and mdm2 may be functionally inhibited at multiple steps to reactivate p53
Red circles indicate potential sites at which small molecule antagonists may be used. First, the interaction of the mdm2 (pink) or mdmx (gold) N terminus with p53 (green) can be antagonized using small molecules such as Nutlin (1 and 2). Second, antagonism of the interaction between the mdm2 and mdmx RING domains (3), may prevent the formation of an optimal E3 ligase complex. Whether such antagonists would interact with mdm2/x monomers, or with heterodimers remains to be determined. Third, the recruitment of E2 conjugating enzyme carrying activated ubiquitin to mdm2/mdmx is a potential target for antagonists (4). Note that each heterodimer has the potential to bind at least 2 p53 molecules, via the N termini of both mdm2 and mdmx. However, no structural data is available for the N terminal regions of mdm2/mdmx heterodimers in complex with p53 and therefore only one p53 molecule is shown for simplicity.
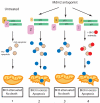
Figure 4. BH3-mediated sensitization to Mdm2 antagonists
In unstressed cells (1), proapoptotic BH3 proteins are attenuated by the activity of antiapoptotic homologs (blue and beige circles respectively). Following antagonism of mdm2, three scenarios can be envisioned. In (2), activation of p53 induces sufficient mdm2 to decrease mdmx levels, and leads to sufficient proapoptotic BH3 activation to induce apoptosis. If mdmx is not degraded or antagonized (3), the amount of p53-induced BH3s may not reach the threshold required for apoptosis. One potential mechanism to abrogate mdmx-dependent protection against apoptosis is to lower the threshold at which BH3 activation will induce apoptosis by titrating antiapoptotic proteins from the system. This can be achieved using BH3 mimetic compounds (red circles, 4).
Similar articles
- Targeting p53-MDM2-MDMX loop for cancer therapy.
Zhang Q, Zeng SX, Lu H. Zhang Q, et al. Subcell Biochem. 2014;85:281-319. doi: 10.1007/978-94-017-9211-0_16. Subcell Biochem. 2014. PMID: 25201201 Free PMC article. Review. - Mdmx and Mdm2: brothers in arms?
Marine JC, Jochemsen AG. Marine JC, et al. Cell Cycle. 2004 Jul;3(7):900-4. Epub 2004 Jul 2. Cell Cycle. 2004. PMID: 15254433 Review. - Stochastic modeling and simulation of the p53-MDM2/MDMX loop.
Cai X, Yuan ZM. Cai X, et al. J Comput Biol. 2009 Jul;16(7):917-33. doi: 10.1089/cmb.2008.0231. J Comput Biol. 2009. PMID: 19580521 Free PMC article. - Regulation of p53: a collaboration between Mdm2 and Mdmx.
Pei D, Zhang Y, Zheng J. Pei D, et al. Oncotarget. 2012 Mar;3(3):228-35. doi: 10.18632/oncotarget.443. Oncotarget. 2012. PMID: 22410433 Free PMC article. Review. - Mdmx promotes genomic instability independent of p53 and Mdm2.
Carrillo AM, Bouska A, Arrate MP, Eischen CM. Carrillo AM, et al. Oncogene. 2015 Feb 12;34(7):846-56. doi: 10.1038/onc.2014.27. Epub 2014 Mar 10. Oncogene. 2015. PMID: 24608433 Free PMC article.
Cited by
- Chemistry and biology of multicomponent reactions.
Dömling A, Wang W, Wang K. Dömling A, et al. Chem Rev. 2012 Jun 13;112(6):3083-135. doi: 10.1021/cr100233r. Epub 2012 Mar 22. Chem Rev. 2012. PMID: 22435608 Free PMC article. Review. No abstract available. - Single-nucleotide polymorphisms in p53 pathway and aggressiveness of prostate cancer in a Caucasian population.
Sun T, Lee GS, Oh WK, Pomerantz M, Yang M, Xie W, Freedman ML, Kantoff PW. Sun T, et al. Clin Cancer Res. 2010 Nov 1;16(21):5244-51. doi: 10.1158/1078-0432.CCR-10-1261. Epub 2010 Sep 20. Clin Cancer Res. 2010. PMID: 20855462 Free PMC article. - Histone deacetylase inhibitors enhance the anticancer activity of nutlin-3 and induce p53 hyperacetylation and downregulation of MDM2 and MDM4 gene expression.
Palani CD, Beck JF, Sonnemann J. Palani CD, et al. Invest New Drugs. 2012 Feb;30(1):25-36. doi: 10.1007/s10637-010-9510-7. Epub 2010 Aug 3. Invest New Drugs. 2012. PMID: 20680659 - A stapled p53 helix overcomes HDMX-mediated suppression of p53.
Bernal F, Wade M, Godes M, Davis TN, Whitehead DG, Kung AL, Wahl GM, Walensky LD. Bernal F, et al. Cancer Cell. 2010 Nov 16;18(5):411-22. doi: 10.1016/j.ccr.2010.10.024. Cancer Cell. 2010. PMID: 21075307 Free PMC article. - Validation of MdmX as a therapeutic target for reactivating p53 in tumors.
Garcia D, Warr MR, Martins CP, Brown Swigart L, Passegué E, Evan GI. Garcia D, et al. Genes Dev. 2011 Aug 15;25(16):1746-57. doi: 10.1101/gad.16722111. Genes Dev. 2011. PMID: 21852537 Free PMC article.
References
- Rayburn E, Zhang R, He J, Wang H. MDM2 and human malignancies: expression, clinical pathology, prognostic markers, and implications for chemotherapy. Curr Cancer Drug Targets. 2005;5:27–41. - PubMed
- Levav-Cohen Y, Haupt S, Haupt Y. Mdm2 in growth signaling and cancer. Growth Factors. 2005;23:183–192. - PubMed
- Harris LC. MDM2 splice variants and their therapeutic implications. Curr Cancer Drug Targets. 2005;5:21–26. - PubMed
- Chene P. Inhibiting the p53-MDM2 interaction: an important target for cancer therapy. Nat Rev Cancer. 2003;3:102–109. - PubMed
- Parant J, Chavez-Reyes A, Little NA, et al. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet. 2001;29:92–95. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous