MicroRNAs: target recognition and regulatory functions - PubMed (original) (raw)
Review
MicroRNAs: target recognition and regulatory functions
David P Bartel. Cell. 2009.
Abstract
MicroRNAs (miRNAs) are endogenous approximately 23 nt RNAs that play important gene-regulatory roles in animals and plants by pairing to the mRNAs of protein-coding genes to direct their posttranscriptional repression. This review outlines the current understanding of miRNA target recognition in animals and discusses the widespread impact of miRNAs on both the expression and evolution of protein-coding genes.
Figures
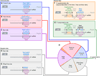
Figure 1. Types of miRNA target sites
(A–C) Canonical, 7–8-nt seed-matched sites. Vertical dashes indicate contiguous Watson–Crick pairing. (D–E) Marginal, 6-nt sites matching the seed region. These 6mer sites typically have reduced efficacy (Figure 4A) and are conserved by chance more frequently than the larger sites. Therefore, when prioritizing site efficacy and prediction specificity, prediction algorithms with stringent seed-pairing criteria disregard 6mer sites. (F–G) Sites with productive 3´ pairing. For 3´-supplementary sites (F), Watson–Crick pairing usually centering miRNA nucleotides 13–16 (orange) supplements a 6–8-nt site (A–E). At least 3–4 well-positioned contiguous pairs are typically required for increased efficacy, which explains why 3´-supplementary sites are atypical. For 3´-compensatory sites (G), Watson–Crick pairing usually centering on miRNA nucleotides 13–16 (orange) can compensate for a seed mismatch and thereby create a functional site. (H) Number of preferentially conserved mammalian sites matching a typical highly conserved miRNA (Friedman et al., 2008). For each site matching the seed region, orange-hatched subsectors indicate the fraction of conserved sites with preferentially conserved 3´-supplementary pairing. Analysis was performed using the 87 miRNA families highly conserved in vertebrates. A 7mer site is counted only if it is not part of an 8mer site, and a 6mer site is counted only if it is not part of a larger site. Values plotted are the number of preferentially conserved sites confidently detected above background, calculated as the average number of conserved sites minus the upper 95% confidence limit on the sites estimated to be conserved by chance. Thus for each site type of panels A–E, there is 95% confidence that the actual average of preferentially conserved sites is higher than that plotted.
Figure 2. Examples of conserved miRNA targets and sites
Coloring of site types and residues within the miRNAs and targets are as in Figure 1. (A) Correspondence between the top predicted targets and genetically implicated interactions (Wightman et al., 1991; Abrahante et al., 2003; Johnston and Hobert, 2003; Lai et al., 2005; Mayr et al., 2007; Xiao et al., 2007). The nematode and mammalian predictions are the top Targetscan (release 4.2) predictions for the respective miRNAs, whereas the fly predictions are among several top but essentially equivalent predictions. Many are also among the top three predictions for the respective miRNAs at the PicTar (lin-14, hbl-1, Brd, Hmga2), EMBL (E(Spl), Brd), and EIMMo (hbl-1, Brd, Hmga2) web sites. Sites within cooperative distance of each other are indicated (square brackets). (B) Additional examples of nematode targets originally identified with assistance from genetics (Moss et al., 1997; Reinhart et al., 2000). (C) Top ORF target candidate of mammalian let-7 (Lewis et al., 2005). (D) Examples of canonical miRNA target sites (Lee et al., 1993; Wightman et al., 1993; Poy et al., 2004; Lai et al., 2005), with sites and pairing reflecting current understanding of target recognition. (E) A 3´-supplementary site with experimental support (Brennecke et al., 2005). (F) 3´-compensatory sites with experimental support (Vella et al., 2004; Yekta et al., 2004). The position of miRNA-directed cleavage within the HoxB8 3´ UTR is indicated (arrowhead).
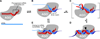
Figure 3. A speculative model explaining the roles of each miRNA region
(A) MicroRNA (red) bound by Argonaute (AGO) such that nucleotides 2–8 are preorganized to favor efficient pairing. Nucleotide 1 is twisted away from the helix and permanently unavailable for pairing; nucleotides 9–11 are facing away from an incoming mRNA and unavailable for nucleation; the remainder of the miRNA is bound in a configuration that has not been preorganized for efficient pairing. (B) Recognition of an 8mer site by the preformed binding pocket. An A at position 1 of the site presumably is recognized directly by the protein of the silencing complex. (C) Massive conformational accommodation of extensively paired sites. At very rare sites of endogenous miRNAs (and the intended sites of exogenous siRNAs) pairing is extensive. In this model, pairing anchored and nucleated at the seed extends to the central region of the miRNA causing the protein to loosen its grip on the 3´ region of the miRNA and thereby allowing the miRNA and mRNA to wrap around each other. (D) Accommodated pairing suitable for mRNA cleavage. The Argonaute protein locks down on the extensively paired duplex, which places the active site (black arrowhead) in position to cleave the mRNA. (E) 3´-supplementary pairing. The message can pair to nucleotides 13–16, incorporating them into a short helical segment without major perturbation of the Argonaute protein or the remainder of the miRNA. Importantly, in this mode of target recognition, the miRNA and mRNA are not wrapped around each other.
Figure 4. Relative mean efficacy of miRNA sites
Efficacy is plotted on a log scale as mean destabilization of messages possessing the indicated sites corresponding to an introduced miRNA, as monitored on mRNA expression arrays (Grimson et al., 2007). Site-conservation analyses, site-depletion analyses, reporter-assay results, and proteomic results all indicate that these differences measured at the mRNA level correspond to similar relative differences in protein output (Grimson et al., 2007; Baek et al., 2008). The four seed-matched sites are colored as in Figure 1. Nonconserved sites are considered together with conserved sites, and unless noted otherwise, only messages with single sites within the 3´ UTR are considered. The _Y_-axis is marked off in units corresponding to the mean efficacy of a single 7mer, considering both types of 7mers in aggregate (dashed lines). (A) Mean efficacy of sites differentiated by their type. Efficacies of the four seed-matched sites (Figure 1A–D) are plotted. To illustrate the contribution of seed pairing, also plotted are efficacies of 8mer sites with a single mismatch or wobble at nucleotides 3, 4, 5, 6 or 7 (open purple bar) and of the offset 6mer match (Figure 1E, open black bar). (B) Mean efficacies of a single 7mer and dual 7mers. Both types of 7mer sites are considered in aggregate (black filled bars). (C) Mean efficacy of sites differentiated based on their position in the message. (D) Mean efficacy of sites differentiated based on their local AU content. (E) Mean efficacy of sites differentiated based on potential for 3´-supplementary pairing.
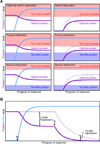
Figure 5. MicroRNA-mediated regulatory effects
(A) Classes of miRNA regulatory interactions (Bartel and Chen, 2004). Left panels: in response to a developmental or environmental cue, cells induce miRNA expression (blue), which in turn dampens protein production (purple) from the targeted message. Right panels: protein production from analogous targets also can be dampened by pre-existing miRNAs; accumulation in the absence of the miRNA is indicated (dashed line). All panels depict 33% repression, which illustrates that classification does not rely on the magnitude of repression but instead depends on the properties of each target, namely its threshold(s) for optimal protein output. These thresholds are depicted as the boundaries between the clear (optimal) and colored (suboptimal) regions of the graphs. (B) Modest destabilization transitions to much greater repression after the mRNA is no longer transcribed. A miRNA is induced (point a) and mediates modest (2-fold) mRNA destabilization. After transcription of the mRNA halts (point b), the same modest destabilization quickly yields substantial (≥10-fold) repression of protein output. If the miRNA also mediates translational repression (not considered in this example), the transition to the off state is accelerated further.
Similar articles
- Small but influential: the role of microRNAs on gene regulatory network and 3'UTR evolution.
Zhang R, Su B. Zhang R, et al. J Genet Genomics. 2009 Jan;36(1):1-6. doi: 10.1016/S1673-8527(09)60001-1. J Genet Genomics. 2009. PMID: 19161940 Review. - Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs.
Lai EC, Tam B, Rubin GM. Lai EC, et al. Genes Dev. 2005 May 1;19(9):1067-80. doi: 10.1101/gad.1291905. Epub 2005 Apr 15. Genes Dev. 2005. PMID: 15833912 Free PMC article. - Biological basis for restriction of microRNA targets to the 3' untranslated region in mammalian mRNAs.
Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Gu S, et al. Nat Struct Mol Biol. 2009 Feb;16(2):144-50. doi: 10.1038/nsmb.1552. Epub 2009 Feb 1. Nat Struct Mol Biol. 2009. PMID: 19182800 Free PMC article. - Spatial preferences of microRNA targets in 3' untranslated regions.
Majoros WH, Ohler U. Majoros WH, et al. BMC Genomics. 2007 Jun 7;8:152. doi: 10.1186/1471-2164-8-152. BMC Genomics. 2007. PMID: 17555584 Free PMC article. - MiR-34a and endothelial biology.
Li Q, Zhang Q. Li Q, et al. Life Sci. 2023 Oct 1;330:121976. doi: 10.1016/j.lfs.2023.121976. Epub 2023 Jul 24. Life Sci. 2023. PMID: 37495076 Review.
Cited by
- Establishment and assessment of an oral squamous cell carcinoma N7-methylguanosine methyltransferase associated microRNA prognostic model.
Li J, Li C, Li X, Chen Y, Li Z, Lin Y, Jing H, Wang Y, Yang H. Li J, et al. J Cancer. 2024 Sep 30;15(18):6022-6037. doi: 10.7150/jca.98350. eCollection 2024. J Cancer. 2024. PMID: 39440068 Free PMC article. - Circulating hsa-miR-323b-3p in Huntington's Disease: A Pilot Study.
Ferraldeschi M, Romano S, Giglio S, Romano C, Morena E, Mechelli R, Annibali V, Ubaldi M, Buscarinu MC, Umeton R, Sani G, Vecchione A, Salvetti M, Ristori G. Ferraldeschi M, et al. Front Neurol. 2021 May 5;12:657973. doi: 10.3389/fneur.2021.657973. eCollection 2021. Front Neurol. 2021. PMID: 34025560 Free PMC article. - Role of microRNAs in the immune system, inflammation and cancer.
Raisch J, Darfeuille-Michaud A, Nguyen HT. Raisch J, et al. World J Gastroenterol. 2013 May 28;19(20):2985-96. doi: 10.3748/wjg.v19.i20.2985. World J Gastroenterol. 2013. PMID: 23716978 Free PMC article. Review. - An SREBP-responsive microRNA operon contributes to a regulatory loop for intracellular lipid homeostasis.
Jeon TI, Esquejo RM, Roqueta-Rivera M, Phelan PE, Moon YA, Govindarajan SS, Esau CC, Osborne TF. Jeon TI, et al. Cell Metab. 2013 Jul 2;18(1):51-61. doi: 10.1016/j.cmet.2013.06.010. Cell Metab. 2013. PMID: 23823476 Free PMC article. - Regulation of cell proliferation and migration in glioblastoma: new therapeutic approach.
Kim Y. Kim Y. Front Oncol. 2013 Mar 18;3:53. doi: 10.3389/fonc.2013.00053. eCollection 2013. Front Oncol. 2013. PMID: 23508546 Free PMC article.
References
- Abelson JF, Kwan KY, O'Roak BJ, Baek DY, Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rasin MR, Gunel M, et al. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science. 2005;310:317–320. - PubMed
- Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4:625–637. - PubMed
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans . Curr Biol. 2003;13:807–818. - PubMed
- Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials