Over half of breakpoints in gene pairs involved in cancer-specific recurrent translocations are mapped to human chromosomal fragile sites - PubMed (original) (raw)
Over half of breakpoints in gene pairs involved in cancer-specific recurrent translocations are mapped to human chromosomal fragile sites
Allison A Burrow et al. BMC Genomics. 2009.
Abstract
Background: Gene rearrangements such as chromosomal translocations have been shown to contribute to cancer development. Human chromosomal fragile sites are regions of the genome especially prone to breakage, and have been implicated in various chromosome abnormalities found in cancer. However, there has been no comprehensive and quantitative examination of the location of fragile sites in relation to all chromosomal aberrations.
Results: Using up-to-date databases containing all cancer-specific recurrent translocations, we have examined 444 unique pairs of genes involved in these translocations to determine the correlation of translocation breakpoints and fragile sites in the gene pairs. We found that over half (52%) of translocation breakpoints in at least one gene of these gene pairs are mapped to fragile sites. Among these, we examined the DNA sequences within and flanking three randomly selected pairs of translocation-prone genes, and found that they exhibit characteristic features of fragile DNA, with frequent AT-rich flexibility islands and the potential of forming highly stable secondary structures.
Conclusion: Our study is the first to examine gene pairs involved in all recurrent chromosomal translocations observed in tumor cells, and to correlate the location of more than half of breakpoints to positions of known fragile sites. These results provide strong evidence to support a causative role for fragile sites in the generation of cancer-specific chromosomal rearrangements.
Figures
Figure 1
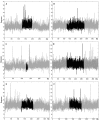
DNA flexibility analysis of translocation-prone and fragile site co-localized genes. DNA sequences within and flanking genes (A) CBFB (B) MYH11 (C) HMGA1 (D) LAMA4 (E) MLL (F) AFF4 were analyzed using the FlexStab program. The analysis was performed over the length of the entire gene (shaded in black) plus 125 kb flanking on each side (shaded in gray). The x axis indicates the size of the analyzed sequences, and the y axis shows degrees of inclination in the twist angle. Windows with values > 13.7° were considered as significantly high flexibility peaks [17].
Figure 2
Secondary structure analysis of CBFB, MYH11, HMGA1, LAMA4, MLL, and AFF4 loci. (A) Comparison of potential to form secondary structure for these genes versus a control. The computed lowest free energy of predicted DNA secondary structures from segments of 300 nt in length, overlapping in 150 nt steps, has been fit to a curve for each gene. The Matlab function polyfit finds coefficients of a polynomial P(X) of degree N that fit the raw data best in a least-squares sense. The analysis was performed over the length of the entire gene plus 125 kb flanking on each side. The arrows indicate where a gene begins and ends. The control sequence was generated by randomizing LAMA4 1000 times. The x axis indicates the size of the analyzed sequences, and the y axis displays the free energy of the predicted structure. Raw data plots for each gene are included in Additional file 3. (B) The most stable structure predicted for each gene, as produced by MFOLD. Each structure represents the 300 nt segment with the lowest ΔG value.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA085826/CA/NCI NIH HHS/United States
- T32 GM095440/GM/NIGMS NIH HHS/United States
- CA85826/CA/NCI NIH HHS/United States
- CA113863/CA/NCI NIH HHS/United States
- R01 CA113863/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources